Figure 5. Antibody 2C06 partially accommodates, but antibody 2C09 clashes with, glycan241.
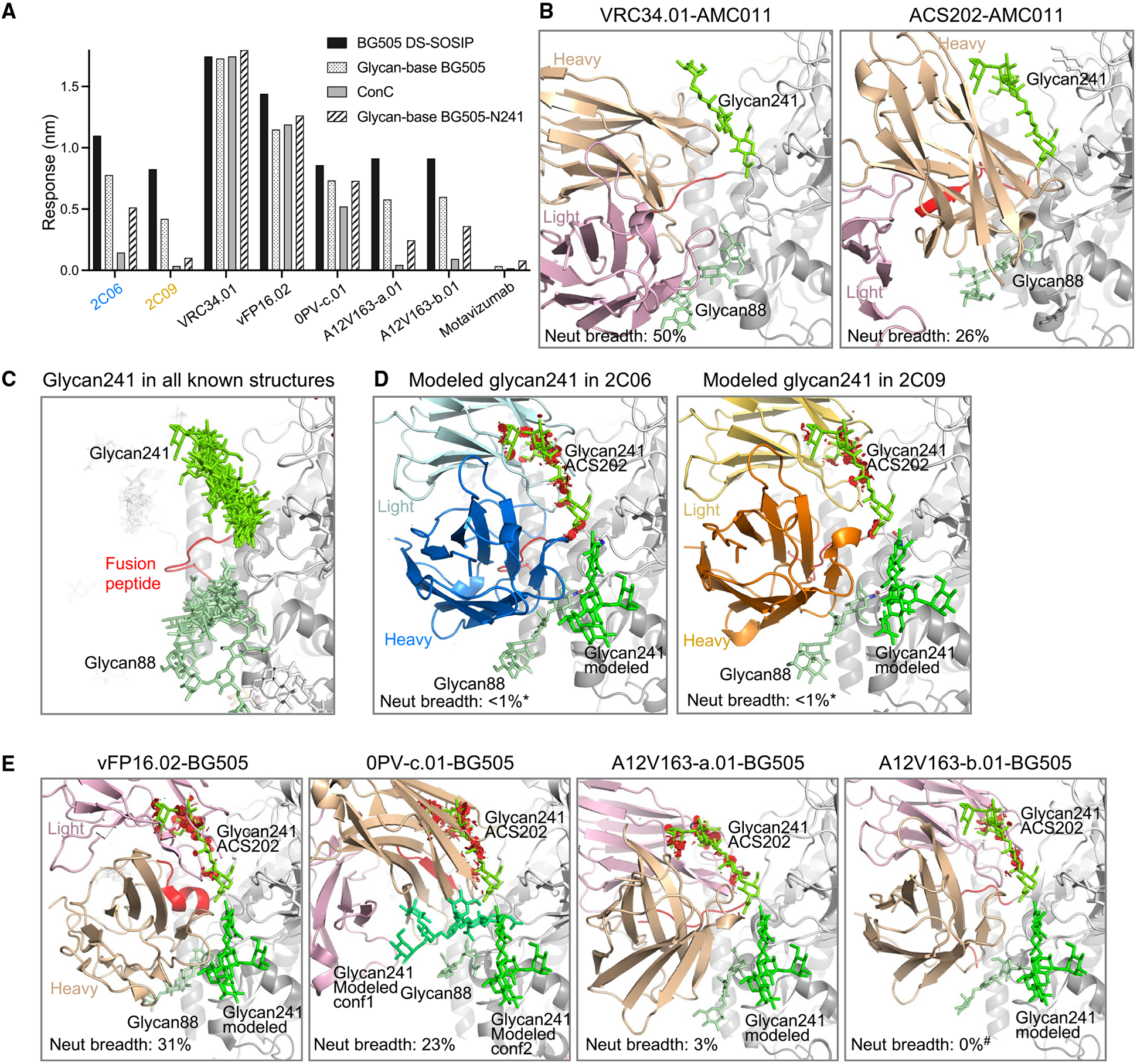
(A) BLI binding analysis of 2C06 and 2C09 with Env trimers with or without glycan241 in comparison with other fusion-peptide-directed antibodies. Both BG505 DS-SOSIP and glycan-base BG505 have serine at 241. Glycan-base BG505-N241 is an S241N mutant of glycan-base BG505 containing N-glycosylation sequon at 241. ConC harbors glycan241. Motavizumab is a negative control. Data were measured once.
(B) Structures of VRC34.01 (PDB: 6nc3) and ACS202 (PDB: 6nc2), both in complex with AMC011 Env, which contains glycan241, reveal that both antibodies interact favorably with glycan88 and glycan241, shown in different shades of green. Neutralization breadths were for half-maximal inhibitory concentration (IC50) < 50 μg/mL in the 208-strain panel.
(C) Structural alignment of Env trimers from the PDB with two or more sugar residues built for glycan241. The structures were aligned by the gp120 subunit bearing the glycan241 shown. Antibodies, if present in the structure, are not shown for clarity. Glycan88 and glycan241 are shown in different shades of green, with the same view as in other structural panels of the figure. PDB: 6olp, 6vo3, 7rsn, 7rso, 6ohy, 7n6u, 6vrw, 6myy, 7llk, 6u59, 5vn8, 6p65, 6vy2, 7lua, 6okp, 7l6o, 6nc3, 6nc2.
(D) Antibodies 2C06 and 2C09 clash with the glycan241 conformation seen in all known Env structures containing glycan241. The structures were aligned with that of ACS202-AMC011 complex by the gp120 subunit bearing the glycan241 shown. Clashes between antibody 2C06 or 2C09 and glycan241 in the ACS202 complex are shown as red disks in PyMOL (https://pymol.org/2/). A different glycan conformation could be modeled (shown in bright green) that did not clash with the antibody. Asterisks denote that 2C06 and 2C09 neutralize BG505 only.
(E) Antibodies derived from mouse immunizations, vFP16.02 (PDB: 6cdi), and from non-human primates, 0PV-c.01 (PDB: 6nf2), A12V163-a.01 (PDB: 6n1v), and A12V163-b.01 (PDB: 6mgp), all of which clash with the glycan241 conformation in ACS202. A different glycan241 conformation, similar to that for 2C06 and 2C09, could be modeled, which avoids any clashes. Binding of 0PV-c.01 allows for more space for multiple conformations of modeled glycan241. Hash mark denotes that A12V163-b.01 neutralizes only at IC50 > 50 mg/mL.
See also Figures S2 and S5; Table S4.
