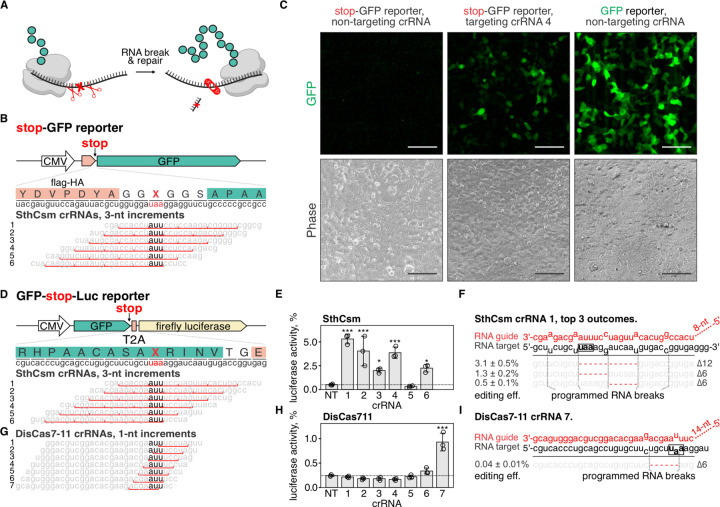
Fig. 3. Programmable deletion of stop codons restores protein expression.
A) Schematic representation of the proposed approach for deleting premature stop codons in human transcripts. B) Top: schematic diagram of the stop-GFP reporter plasmid. Bottom: Six guide RNAs for SthCsm complex were designed to excise the stop codon in the gfp transcript. Underlined (red) sections of target RNA are expected to be deleted. Vertical red ticks indicate predicted sites for RNA breaks. C) Cells were transfected with plasmids for the stop-GFP reporter and SthCsm with a non-targeting guide (left), stop-GFP reporter and SthCsm with a targeting guide (middle), or GFP reporter and SthCsm with the non-targeting guide (right). Fluorescence microscopy was used to image cells 48 h post-transfection. D) Top: schematic diagram of the GFP-stop-Luc reporter plasmid. Bottom: Six crRNAs for SthCsm complex were designed to delete the stop codon at the 3’-end of the gfp gene. E) Luciferase activity was measured in cell lysates 48 hours after transfection with GFP-stop-Luc and SthCsm plasmids. Luciferase activity is normalized to a control transfected with a reporter plasmid without the stop codon. Data are shown as mean ± one standard deviation of three replicates. Means were compared using one-way ANOVA, and samples with targeting guide RNAs were compared to the non-targeting control using one-tailed Dunnett’s test. * p < 0.5, ** p < 0.1, *** p < 0.001. F) Most frequent RNA editing outcomes in the sample with the most efficient rescue of luciferase activity in (B) (guide 1). Editing efficiency was quantified as mean ± one standard deviation of three biological replicates. Black box shows the stop codon that was targeted by type III CRISPR complexes. G, H, I). The same as (D-F), but with DisCas7-11.