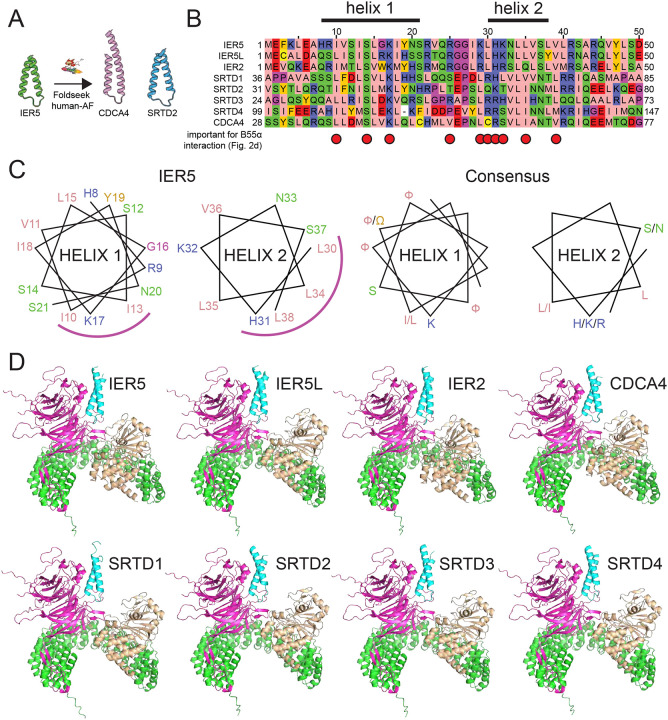
Fig. 5. Identification of sequence and structural homology between IER family members and SERTA domain containing proteins.
A, Searching the human alphafold predicted proteome using IER5 as input in Foldseek28 led to the identification of CDCA4 and SRTD2 as structural homologues. B, Multiple sequence alignment focusing on the helix-loop-helix motif of IER5, aligning IER, SRTAD, and CDCA4 proteins. The helix 1 and helix 2 segments of IER5 seen in the structure of the PP2A/B55α complex with IER5-N50 are indicated above the alignment. Red dots indicate sites of mutations in IER5 that interfere with co-immunoprecipitation of B55α. The alignment is shown using the Zappo color scheme for the 20 amino acids: pink, aliphatic; green, hydrophilic; blue, basic; red, acidic; orange, aromatic; yellow, cysteine; magenta, proline or glycine. C, Helical wheel diagram of IER5 helix 1 and helix 2 (left) and a consensus for helix 1 and 2 based on residue conservation among aligned IER, SRTAD and CDCA4 proteins (aliphatic and aromatic residues are denoted as F or W, respectively). The helical face directed at B55α is marked by a magenta arc. D, Best scoring structural models for alphafold2-predicted interactions of IER, SERTAD and CDCA4 proteins with PP2A/B55α. IER, SERTAD, and CDCA4 predictions were restricted to the aligned region in (B). B55α is magenta, the PP2A catalytic subunit is wheat, the PP2A scaffolding subunit is green, and the predicted interactor is cyan. See also Table S1.