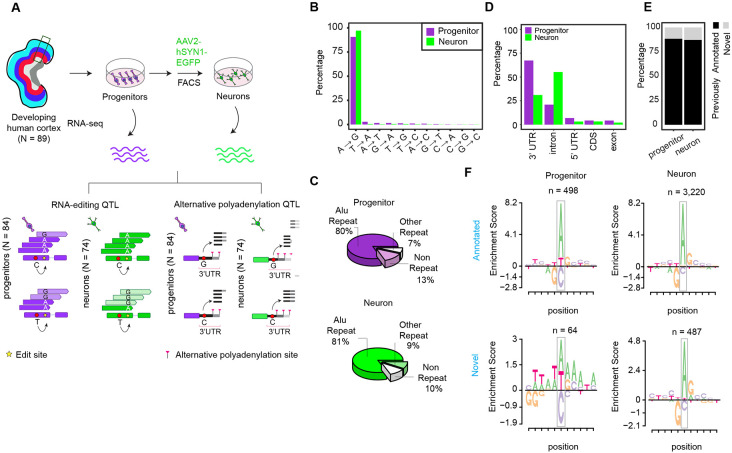
Figure 1. Study design and features of RNA-editing sites.
(A) Study design to identify cell-type-specific edQTLs and apaQTLs.
(B) Proportions of RNA-editing events discovered in each cell type.
(C) Pie chart showing proportions of RNA-editing events within Alu repeats, other repeats or non-repeat regions in the genome per cell type.
(D) Proportions of RNA-editing events across genomic regions. CDS: Coding sequence, 3’UTR: Three prime untranslated region and 5’UTR: Five prime untranslated region
(E) Overlap of RNA-edit sites with GTEx Cortex or BrainVar datasets. The edit sites overlapped are defined as annotated and the sites which do not overlap are defined as novel.
(F) Local motif enrichment for annotated and novel edit sites per cell-type. Number of edit sites (n) in each category is reported.