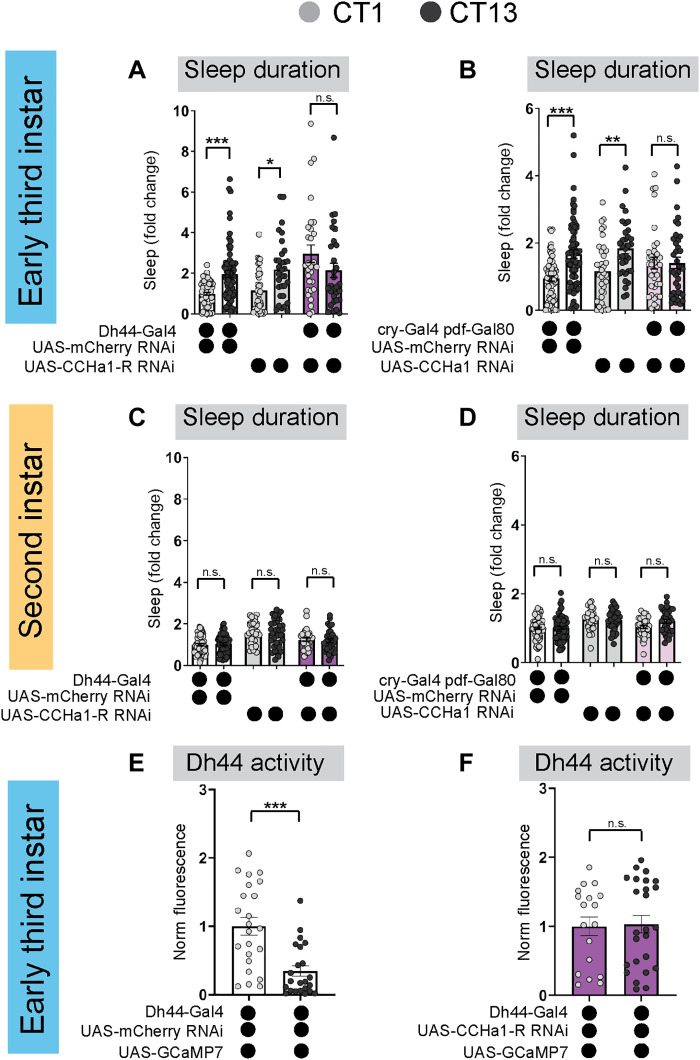
Fig. 4. CCHa1 signaling between DN1a and Dh44 neurons is necessary for sleep rhythms.
(A and B) Sleep duration in third-instar (L3) expressing UAS-CCHa1-R-RNAi with Dh44-Gal4 (A) and UAS-CCHa1-RNAi with cry-Gal4 pdf-Gal80 (DN1as) (B) and genetic controls at circadian time 1 (CT1) and CT13. (C and D) Sleep duration in second-instar (L2) expressing UAS-CCHa1-R-RNAi with Dh44-Gal4 (C) and UAS-CCHa1-RNAi with cry-Gal4 pdf-Gal80 (DN1as) (D) and genetic controls at CT1 and CT13. (E and F) GCaMP intensity (normalized fluorescence) in Dh44 neurons in L3 expressing UAS-CCHa1-R-RNAi with Dh44-Gal4 and genetic controls at CT1 and CT13. Sleep metrics represent fold change (normalized to the average value of control). (A to D) n = 64 to 68 mCherry larvae, 29 to 37 other genotypes; (E and F) n = 18 to 25 cells, 10 brains. Two-way analyses of variance (ANOVAs) followed by Sidak’s multiple comparisons tests (A to D); unpaired two-tailed Student’s t test (E and F). n.s., not significant.