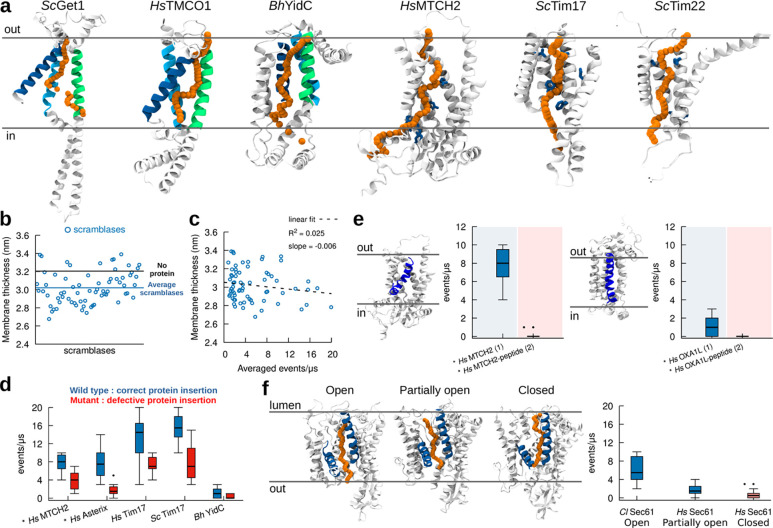
Figure 4. Lipid scrambling takes place via the same mechanistic pathway as in protein insertion.
A. In silico lipid scrambling pathway (orange) in selected protein insertases. The position of the lipid polar head at different times along the scrambling pathway is depicted with orange spheres. Regions involved in protein insertion are shown in green, blue and cyan for Oxa1 family proteins, while residues involved in protein insertion are shown in blue for HsMTCH2, ScTim17 and ScTim22. B. Protein scramblases induce limited (0.2 nm on average) membrane thinning. C. Membrane thickness has minimal correlation with lipid scrambling activity in silico. D. Mutants proposed to decrease protein insertion activity also reduce lipid scrambling. E. The presence of a nascent polypeptide inside the protein hydrophilic cavity abolishes lipid scrambling. Left: HsMCTH2. Right: HsOXA1L. f. Different conformations of Sec61 (lateral gate open, partially open and closed) have different lipid scrambling activity. AlphaFold structures are denoted by the * symbol. Number of proteins in the system is in parenthesis.