Abstract
Throughout the history of electron microscopy, ribosomes have served as an ideal subject for imaging and technological development, which in turn has driven our understanding of ribosomal biology. Here, we provide a historical perspective at the intersection of electron microscopy technology development and ribosome biology and reflect on how this technique has shed light on each stage of the life cycle of this dynamic macromolecular machine. With an emphasis on prokaryotic systems, we specifically describe how pairing cryo-EM with thoughtful experimental design, time-resolved techniques, and next-generation heterogeneous structural analysis has afforded insights into the modular nature of assembly, the roles of the many transient biogenesis and translation co-factors, and the subtle variations in structure and function between strains and species. This work concludes with a prospective outlook on the field, highlighting the pivotal role cryogenic electron tomography is playing in adding cellular context to our understanding of ribosomal life cycles, and noting how this exciting technology promises to bridge the gap between cellular and structural biology.
Ribosomes and electron microscopy: a history intertwined.
The ribosome, which is responsible for protein translation in all domains of life, is a massive ribo-nucleoprotein complex ranging in size from ~2.5 MDa in bacteria1 to ~4 MDa in eukaryotes2. This essential molecular machine is comprised of two subunits formed by interwoven RNA helices and ribosomal proteins that assemble through a dynamic, multi-step biogenesis pathway3,4. Once assembled, the machine is itself dynamic, undergoing structural changes coupled to its function in mRNA binding and decoding, peptidyl transferase activity, and its eventual sequestration and degradation. These dynamics involve both conformational motions and regulated compositional changes, with more than 50 core proteins and hundreds of transient binders and cofactors associating with the particle during its assembly and as it functions5.
Electron microscopy (EM), including negative stain EM, single particle cryogenic EM, and cryogenic electron tomography, have proven to be uniquely suited for studying the ribosome. Unlike X-ray crystallography, in which a requisite crystal lattice locks the ribosome in a single structural state, cryoEM observes 104–107 individual and, potentially, structurally heterogeneous particles in a single experiment. Although such structural heterogeneity presents computational challenges for the reconstruction and refinement of cryoEM density maps, it also provides an opportunity to see the full range of structural states populated in a sample6. With the advent of improved imaging technology7,8 and 3D-reconstruction algorithms9–11, ribosome structural dynamics are being systematically characterized in greater detail than ever before12 (Fig. 1).
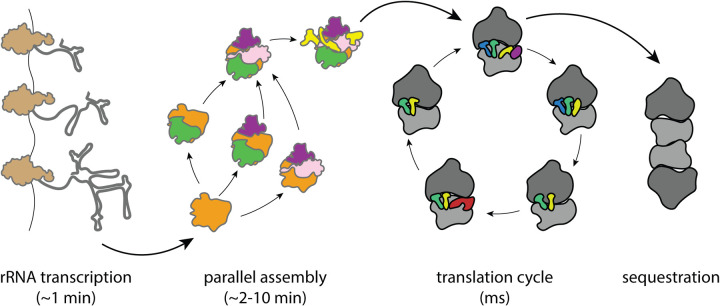
Figure 1. Selected phases from the bacterial ribosome lifecycle.
The ribosome lifecycle involves an array of heterogeneous structural states related by both parallel and sequential pathways across timescales spanning several orders of magnitude.
Early studies of the ribosome are inextricably linked with the development of electron microscopy. These “small particulate component[s] of the cytoplasm” that were briefly known as “particles of Palade,” were first identified in 1955 in electron micrographs of the endoplasmic reticulum of rat liver cells13,14, though Salvador Luria had incidentally imaged ribosomes in his electron micrographs of bacteriophages years prior15. Early evidence for co-transcriptional ribosome assembly, which is discussed below, came from direct visualization of rRNA synthesis in rapidly growing cells using “Miller spreads” – a method that gently disperses chromatin and fixes RNA polymerases to their DNA templates and nascent transcripts16–18. Negative stain electron micrographs of such Miller spreads revealed dense packing of RNA polymerases along E. coli rRNA operons, with fibrils of increasing lengths emanating from the RNA polymerase. These fibrils were decorated with protein-dense ribosome assembly intermediates, which collectively gave rise to a “Christmas tree” morphology (Fig. 2) that highlighted the co-transcriptional nature of ribosome assembly.
Figure 2. Early studies of ribosomes and electron microscopy.
(a) Electron micrograph of endoplasmic reticulum from rat pancreatic acinar cells, first published13 by George Palade in 1954. g – Palade’s “granules” i.e. ribosomes, n – nucleus, x – regions where granules appear to be attached to the membrane of the endoplasmic reticulum, nm1 – nuclear membrane proper, nm2 – endoplasmic reticulum membrane limiting the cytoplasm toward the nucleus, z – zymogen granule13. (b) Electron micrograph of a gene transcription unit prepared by the Miller spreading technique158. (c) The first 3-dimensional models of the 30S (left) and 50S (right) subunits, with single particle images from electron micrographs in corresponding orientations20. Figure panels reproduced with permission from above references.
Such applications of electron microscopy not only advanced the field’s understanding of ribosomes, but many were also critical in advancing electron microscopy as a technology. Indeed, the large, dense, and abundant nature of ribosomes make them nearly ideal imaging subjects19 and, as such, they have played key roles in the development of negative stain and cryogenic EM20,21, image processing and 3D-reconstruction methods21–25, the development of early heterogeneous classification and reconstruction algorithms26, and the more recent development of cryoEM and cryo-electron tomographic methods to determine structures in situ27–30. Likewise, some of the earliest examples of exhaustive structure classification methods aimed at understanding function31,32, and the development of methods to model continuous forms of heterogeneity were tested using datasets of ribosome assembly intermediates that exhibited vast structural heterogeneity6,11,12,33,34.
In this chapter, we highlight how methodological advances in electron microscopy have enabled a greater understanding of the structural dynamics occurring in ribosomes as they undergo assembly and as they function, providing vignettes where electron microscopy has played a central role. Although EM has contributed to our understanding of ribosome structure-function relationships in both eukaryotes and prokaryotes, we focus on the latter, and direct readers to excellent recent reviews of analogous studies of eukaryotes35,36.
Snapshots of ribosome biogenesis.
Ribosome biogenesis is a rapid, multi-step process, involving the interplay of ribosomal proteins (r-proteins) and RNA (rRNA), as well as transiently bound assembly cofactor proteins. In prokaryotic cells, much of the particle’s assembly occurs co-transcriptionally, with rRNA folding coupled to r-protein binding events that help to stabilize productively folded rRNA, thereby guiding the particle’s maturation4,18,37–39. Notably, however, pioneering work by the Nomura and Nierhaus groups that reconstituted assembly of the 30S and 50S subunits in vitro showed that purified, fully transcribed rRNAs and r-proteins can self-assemble into active ribosomal subunits, which argued that co-transcriptional assembly is not strictly required for ribosome biogenesis40–49. Moreover, this in vitro approach allowed them to test the binding interdependence between r-proteins through order-of-addition experiments, and these studies provided evidence for thermodynamic coupling between many r-protein binding events. Certain proteins, termed primary binders, could bind the rRNA directly, whereas others, termed secondary and tertiary binders, exhibited stronger binding when other r-proteins were present. These experiments supported a model of hierarchical ribosome subunit assembly involving sequential binding of secondary and tertiary proteins, and parallel assembly of the primary binders.
Whereas these in vitro assembly reactions demonstrated the ability of subunits to assemble using only core ribosomal components, assembly is known to occur more efficiently and more rapidly in cells. Although the co-transcriptional nature of such cellular assembly is likely a dominant contributor to this improvement, a wide array of cellular assembly cofactors, including GTPases, rRNA and r-protein modification enzymes, and RNA helicases are also posited to contribute to the improved assembly efficacy observed in cells4,38,50. Additionally, many of these cofactors profoundly impact assembly efficacy under stress conditions, including cold-shock, suggesting roles for assembly cofactors in overcoming stress-related barriers to productive assembly51. Despite this broad understanding of assembly cofactor roles, there remain several areas of active inquiry, including: when in the assembly process do cofactors act; how do cofactors interact with each other; is there redundancy amongst cofactors; and how do cellular stresses impact the role of these cofactors? Moreover, basic questions about the mechanisms by which these proteins facilitate assembly remain unanswered, with postulated functions including: ensuring the correct assembly order52–54; expediting specific structural changes55,56; and proofreading specific assembly defects12.
Answering these and related questions has proven challenging for three key reasons. First, despite its large size and complexity, the entire E. coli ribosome biogenesis process is completed in roughly two minutes and ribosomal precursors account for only 2–5% of all ribosomes during rapid growth57,58. As a result, isolating and studying specific ribosomal intermediates is difficult. Second, assembly cofactors are believed to associate transiently with their ribosomal substrates, and these substrates are of low abundance in the cell, making them difficult to isolate natively. Finally, multiple parallel assembly pathways exist that produce highly heterogeneous ensembles of assembly intermediates3,31. Approaches to isolate the relatively short-lived intermediates include: genetic systems to perturb r-protein and cofactor levels and thus stall the assembly process31,53,59–62; in vitro reconstitutions, which are inherently slow63–65; and biochemical approaches, such as affinity enrichment of assembly-factor bound particles66. Over the past 10 years, each of these strategies have been coupled to single particle cryoEM to characterize the structure and composition of highly heterogenous populations and, in some instances, to track dynamic structural re-arrangements that occur during assembly. Here, we highlight a non-exhaustive set of such studies that leveraged single particle cryoEM to understand the biogenesis process.
CryoEM reveals conserved structural blocks in ribosome biogenesis.
Whereas the Nierhaus experiments provided a roadmap to understand the consequences of withholding an r-protein from an in vitro large subunit assembly reaction, it was unclear how cellular assembly pathways, which are under exquisite transcriptional and translational control, and bear an array of potentially buffering assembly cofactors, would respond to such a perturbation. To answer this question, Davis et al. depleted the early binding r-protein bL17 from cells, resulting in the accumulation of large subunit intermediates exhibiting both compositional and conformational heterogeneity. These intermediates were biochemically purified and analyzed using quantitative mass spectrometry and cryoEM31 – a form of ex vivo analysis. In one of the first applications of deep structural classification to ribosome biogenesis intermediates, this work revealed 13 distinct intermediate structures resolved at 4–5 Å resolution that allowed for comparison of the presence/absence of each rRNA helix and r-protein (quantified as “occupancy”) across the entire range of ribosomal precursors. This quantification produced an “occupancy map” that, upon hierarchically clustering, revealed sets of similar assembly intermediate particles and sets of rRNA helices and r-proteins whose occupancy profiles were correlated across the 13 structures. Guided by these occupancy maps, the structures were then arranged into an assembly pathway that exhibited hierarchical and parallel elements, and highlighted the malleability of bacterial ribosome assembly. For example, some r-proteins that were known to bind after bL17 in wild-type cells67 were bound to these bL17-free particles, indicating that the assembly order could be permuted when limited quantities of an early binding r-protein were available. Moreover, this approach identified five major groups of rRNA and protein “blocks” that exhibited highly correlated behavior across the accumulating intermediates and co-localized on the mature large subunit. The authors suggested that this modular block-like assembly was a result of thermodynamic cooperativity built into the assembly process, and the subsequent observation of similar assembly blocks in independently analyzed large subunit assembly intermediates is consistent with this notion3,64. Finally, by identifying mutually exclusive blocks among ribosome precursors, which the authors posited would not disassemble once formed, this work provided the first structural evidence of parallel, branched assembly pathways (Fig. 3). Notably, more recent applications of advanced classification and reconstruction approaches, including deep hierarchical classification32 and continuous models of structural heterogeneity have recapitulated these original analyses6,11,33,34.
Figure 3. Parallel assembly pathways observed under bL17 depletion.
The most immature observed class B is shown at the top, which is hypothesized to transition to the most mature class E5 via several branched pathways (black arrows). Incorporated r-proteins are indicated at each transition step. Adapted from Davis et. al 201731.
This study additionally allowed for expansion of the classic Nierhaus assembly map under perturbed ex vivo conditions to include rRNA helices and r-protein binding interdependencies, and provided insights into how different assembly pathways can permute the order and coupling of these r-protein binding and rRNA docking events. Taken together, the flexibility observed in the assembly pathways in this work further highlighted the value of interrogating assembly in vivo. Indeed, we and others have speculated that such flexibility may be an essential and evolutionarily selected feature of bacterial ribosome assembly as it would allow assembly to proceed in the face of transient shortages in particular r-proteins or assembly cofactors, or when cells encounter environmental conditions that disfavor a particular assembly pathway3,68,69.
More recently, the Spahn and Nikolay groups have worked to build a structural understanding of the in vitro reconstituted assembly process pioneered by the Nierhaus group63,64. Importantly, these experiments detail progressive maturation pathways based only on the inherent biochemical properties of rRNAs and r-proteins, effectively insulating the observations from the complications that could arise from interactions with assembly cofactors or regulatory cascades in cells. Interestingly, the overarching themes observed by ex vivo Davis et al., including multiple parallel routes of assembly, were reinforced by these in vitro studies. Moreover, both the in vitro and ex vivo, ribosomal precursors appeared to mature in a domain-wise manner, with early precursors comprised of a stable core lacking a functional peptidyl transferase center and key intersubunit bridging rRNA helices, and a significant fraction of particles lacking a well resolved central protuberance. Of note, this reconstitution approach using purified components allowed Qin and colleagues to catalog one of the least mature assembly intermediates to-date, which may represent the minimal stably folded structural domain64. Interestingly, similar early assembly intermediates were recently discovered both in a sample isolated from an in vitro co-transcriptional assembly reaction65 and that purified from cells lacking the helicase DeaD that were grown at low temperature70, indicating such early assembly states can also be observed during co-transcriptional assembly in vitro and in particles isolated from cells.
A new perspective on biogenesis cofactors.
As above, cryoEM has significantly impacted our understanding of how ribosome biogenesis cofactors affect the assembly process, with most structural studies characterizing the ribosomal particles that accumulate in the absence of a cofactor of interest, or in determining the structure of the assembly-cofactor bound complex. To date, studies have investigated the 30S cofactors RsgA/YjeQ71–76, RbfA72,77, RimM59,73, RimP55, KsgA12,78–80, and Era81,82, and the 50S cofactors RbgA52,53,60,83, YphC/EngB and YsxC61, EngA84, RrmJ85, ObgE66,86, SrmB62, and DeaD70. Here, we highlight the biogenesis cofactors RbgA and KsgA, detailing how cryoEM and genetic perturbations to deplete cells of these cofactors have been applied to interrogate each cofactor’s mechanism of action.
Notably, given the aforementioned flexibility and inherent permutability of the in vivo assembly pathways, interpreting such cofactor depletion experiments is challenging3. Indeed, parallel assembly branches raise the possibility that the accumulating particles one observes upon cofactor depletion are not true substrates for the cofactor but rather particles that have progressed towards a thermodynamically stable state that is no longer able to interact with the factor3,87. Indeed, it is even formally possible that such accumulated particles are not competent to mature, and are therefore by definition off-pathway.
In analyzing the particles that accumulate in the absence of the ribosome biogenesis-associated GTPase RbgA, Jomaa et al. considered these possibilities and, by coupling pulse-labeling to quantitative mass spectrometry, they determined that the majority of the RbgA-depletion particles were in fact competent to mature to functional 70S ribosomes53,57. Interestingly, they further noted that the functional core (i.e. the peptidyl transferase center, PTC) in these particles was highly disordered, despite observing that the majority of the other structural elements have adopted their mature conformations. Whereas the general observation that functional domains mature late predated this work, the effect was even more pronounced in these53,60 and related61 particles that accumulated upon co-factor depletion. This observation has been interpreted as a quality control mechanism inherent in the assembly process that prevents premature particles from engaging in translation until the particle is completely assembled53 – akin to issuing homeowners keys only once the house is fully constructed, with assembly cofactors acting as building inspectors to ensure proper assembly order is maintained and particle maturation is complete before certification.
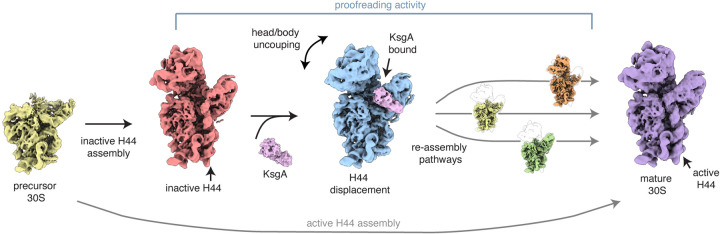
Another method for determining cofactor function is to add the protein of interest back to ribosomes purified from a cofactor-depleted strain and to then assess the changes that occur in vitro. Sun and colleagues employed this strategy while studying the function of KsgA12, a methyltransferase which has long been suspected as a biogenesis cofactor involved in 30S subunit maturation88,89. The group characterized the ensemble of intermediates that accumulate in a cold-sensitive ΔksgA strain of E. coli grown at low temperature, where these accumulating particles were presumed to be KsgA substrates. To assess the impact of KsgA addition to these substrates, they also analyzed structures resulting from addition of purified KsgA to this sample. To their surprise, the KsgA-treated structures were grossly less mature than the particles isolated from the depletion strain, with large swaths of poorly-resolved density in the head, platform and spur domains, indicating greater conformational or compositional heterogeneity in these regions. This led to a hypothesis that KsgA may recognize a structural defect in its substrates and induce disassembly of such particles. Using cryoDRGN6,11 to analyze particles not yet treated with KsgA, they observed that a key helix (H44) could adopt two alternative conformations – one active and the other inactive. Quantitation of the relative abundance of these conformations revealed that upon treatment, KsgA preferentially depleted the sample of H44-inactive particles. Taken together, the evidence suggested that KsgA specifically recognizes inactive 30S intermediates and, through its binding, induces large-scale structural remodeling and disassembly that may afford these inactive particles an opportunity to re-fold and progress down a productive assembly pathway (Fig. 4). In sum, this recognition and disassembly activity was interpreted as KsgA-dependent kinetic proofreading90 of the assembly process. In addition to illuminating a new proofreading role for KsgA, this work also highlighted the potential of next-generation cryoEM analysis tools such as cryoDRGN for analyzing large datasets bearing extensive structural variation, and in using these data to inform our understanding of structural signaling, remodeling, and dynamics.
Figure 4. KsgA proofreading model.
During 30S assembly, immature particles with inactive H44 are recognized by KsgA, which induces subsequent uncoupling of the head/body to provide another opportunity for subunit maturation. Adapted from Sun and Kinman, et al. 202212.
Despite rapid recent progress in understanding bacterial ribosome biogenesis from a structural perspective, many key questions remain. For example, what is the relative flux through different assembly pathways; how do pathways change under different environmental conditions or cofactor depletions; and do multiple cofactors work in concert on a given particle, similar that observed in eukaryotic ribosome assembly intermediates? We expect that sophisticated biochemical and structural analysis protocols will be necessary to capture these more transient stages of assembly and, as techniques improve, we anticipate drawing deeper insights into both the immature assembly intermediates and the mature ribosome as it engages in its primary function in the cell — translation.
A window into prokaryotic translation.
The structural dynamics of translation hold two major themes: compositional dynamics, with tRNAs and GTPase initiation and elongation factors binding and dissociating from the translating ribosome; and conformational dynamics, wherein the subunits undergo movements linked to function91,92. For example, during translation elongation, the large (LSU) and small (SSU) subunits rock back and forth relative to each other in a distinct ratcheting motion that is critical to translocating tRNAs through the active sites93–95, as well as a subsequent step where the SSU head rotates relative to its body in order to move along the mRNA by one codon96.
Due to limitations in microscope and detector hardware, early cryoEM structures of translating ribosomes were relatively low resolution94,97–99. Despite this, targeted studies based on previous biochemical and biophysical work100–103 assembled desired complexes in vitro or stalled the translation complex pharmacologically or genetically, resulting in structures that significantly advanced our understanding of the mechanism and dynamics of translation. Such approaches provided the first visualization of ribosome-bound EF-G, a GTPase that catalyzes mRNA and tRNA translocation during polypeptide elongation104; facilitated discovery of the ratchet motion between the small and large subunits that occurs upon binding of EF-G95; and provided structures of tRNA in hybrid state between the P- and E-sites that occurs as a prelude to tRNA translocation105. Early structures also complemented crystallographic approaches and led to a general understanding that the factors EF-Tu, EF-G, RF-3, and IF-2 bind in the flexible GTPase Associated Center (GAP) via an “induced fit” model, wherein each subsequent factor acts to stabilize the ribosome and promote productive conformational changes to advance the translation cycle97,104,106–108.
Over the past 15 years, technical advances in cryoEM7,109 have produced ever-higher resolution structures that led to new insights into ribosome structure and function. For example, the sub-3 Å resolution structure of a ribosome•EF-Tu complex bound to aminoacyl-tRNA and the antibiotic kirromycin resolved all 35 rRNA modifications110, providing a structural framework to understand antibiotic action and resistance110–112, and it paved the way for a recent study determining 17 high-resolution structures of antibiotic-ribosome co-complexes113. Further methodological improvements culminated in a 1.6 Å resolution structure of a translating ribosome that, incredibly, allowed for direct structure-guided sequencing of the rRNA114. These structures additionally visualized the flexible and enigmatic protein bL9 in a closed conformation and identified residues on bL9 (Glu87) and uS6 (Arg24) that stabilized this closed conformation via a salt bridge. Notably, bL9 is suspected to play a role in fidelity of translation115, frameshifting116,117, polysome formation118, and quality control pathways in collided ribosomes119,120, and the exchange between the extended conformation originally observed in crystal structures at a lattice interface121–123 and the closed conformations observed by Fischer et al. and others may help to explain these multi-modal functions.
Similarly, image analysis tools aimed at handling structural heterogeneity have begun to link mechanism to the structural ensembles within cryoEM datasets that can be computationally resolved. Loveland et al. used such approaches to resolve six near-atomic-resolution ribosome•tRNA•EF-Tu decoding complexes, bearing cognate and near-cognate tRNA-mRNA pairs124. These structures established that discrimination between cognate and near-cognate tRNAs hinges on a single base on the shoulder of the 30S subunit (G530), which can assume an “ON” conformation when properly stabilized by a network of hydrogen bonds between the mRNA and tRNA bases. They further showed that movement into the “ON” conformation aids in EF-Tu activation that precipitates GTP hydrolysis and EF-Tu dissociation from the complex, which represents the final step of aminoacyl-tRNA accommodation into the LSU tRNA A-site. The ability to visualize several related complexes in such detail was critical in elucidating the mechanisms of translational fidelity, and highlighted the value of accurate particle classification algorithms to not only improve resolution, but to also reveal the structural underpinnings of central biological phenomenon.
Whereas the combination of biochemical data and the targeted enrichment of structural states using genetic or pharmacological approaches has allowed researchers to infer the sequence of structural progression3, the field has long-sought to directly determine time-dependent structural ensembles125 and to visualize transitory structural states within these datasets. Pursuing these goals, Carbone et al. analyzed translational elongation as a function of time after the addition of EF-G using standard vitrification equipment126. Despite the relatively poor temporal resolution afforded by such plunge-freezing post-mixing, their intermediate timepoints captured transient translation states with EF-G bound, and a comparison of their structures highlighted the progression of structural states during EF-G-mediated translocation (Fig. 5). They noted that EF-G’s extended conformation both pre-translocation (Structure III) and nearly post-translocation (Structure IV) was consistent with a model of EF-G acting as a rigid pawl, as opposed to a flexible motor that is actively driving translocation through GTP hydrolysis. They concluded that the motion driving translocation, therefore, is inherent and spontaneous internal movement of the ribosome, including the 30S body rotation and head swivel. The study also captured a structure (Structure VI) that did not have EF-G bound, but which was exclusively present in t=24 seconds (s) dataset, indicating that it was a transient state formed directly after EF-G dissociation. The structure showed the 30S head still in the swiveled position, indicating that the reversal of the head swivel occurs after EF-G dissociation.
Figure 5. Time-resolved cryoEM experiments reveal new structures at different steps of translation.
(a) 30S Initiation Complex (IC) particles were combined with 50S particles using a mixing-spraying time resolved cryoEM set-up, resulting in four timepoints spanning 0–600 milliseconds (ms). The population of 70SIC particles initially increased as the 50S particles bound the 30IC particles, before the 70SIC particles converted to 70S Elongation Complexes (70SEC). Adapted from Kaledhonkar et al. 2019129. (b) EF-G was mixed with pre-translocation 70S ribosomes loaded with tRNAfMet in the P site and dipeptidyl-tRNAPro in the A site. Grids were blotted at 0, 24, and 600 seconds (s). The structures show a full progression of the translocation cycle, and include many tRNA hybrid states. Structures III, IV, V, and VI were found only in the 24 s sample. Structure V shows EF-G after domains I and II have partially dissociated. Adapted from Carbone et al. 2021126. (c) Using a rapid-mixing cryoEM set-up, UAA-programmed release complex, (RC0) with tripeptidyl-tRNA in the P site was added to RF1. At 24 ms, roughly 30% of complexes were bound to RF1 in its compact form, while at 60 ms more than 90% of RF1 had converted to its extended conformation. Adapted from Fu et al. 2019130.
Building on the promise of time-resolved cryoEM, recent innovations including the development of fast-mixing microfluidic vitrification instruments127,128 have enabled millisecond-resolved studies of translation initiation and termination129,130. Kaledhonkar et al. used a rapid mixing approach to study the later steps in bacterial translation initiation, mixing 30S Initiation Complexes (IC) with 50S particles to investigate mechanisms of 70S IC formation and its subsequent maturation into the 70S Elongation Complex (EC). Using a custom-built microfluidic device for mixing and rapid vitrification, they collected datasets spanning 20–600 milliseconds (ms) post-mixing. They found that the 70S IC complex abundance peaked near 80 ms, representing ~40% of the particles, and that by the end of the 600 ms more than 60% of the particles were fully converted to 70S EC complexes (Fig. 5a). Notably, this rapid mixing approach was vital in resolving the short-lived 70 IC complex in the absence of perturbations, and helped to distinguish on-pathway complexes from off-pathway intermediates that could result from such perturbations. Inspection of this native 70S IC structure revealed that IF-1 had dissociated, which alleviated a steric clash with H69 of the large subunit and allowed the formation of inter-subunit bridges required for translation. Moreover, in the transition from the 70S IC to the 70S EC, the authors observed dissociation of IF-2, which appeared to trigger a reverse rotation of the small subunit by 3Å, thereby stabilizing the non-rotated inter-subunit orientation. Finally, the combination of this rotation and the absence of IF-2 enabled the fMet-tRNA to move from the peptidyl/initiation (P/I) configuration observed in the 70S IC to the peptidyl-peptidyl (P/P) configuration observed in the 70S EC, effectively placing the fMet moiety in a peptidyl-transfer competent position within the P-site, and enabling the complex to proceed with the first round of translation elongation.
At the other end of translation, Fu et al. used a similar technique to study the structural changes in release factors (RFs) during translation termination130. RFs contain structural motifs involved in recognizing a stop codon, as well as a GGQ motif that is responsible for ester bond hydrolysis, which allows the nascent peptide release from the ribosome. Interestingly, crystal structures of isolated bacterial RFs are compact, with only 20 Å between the stop codon recognition and GGQ motifs, whereas ribosome-bound RFs adopt an extended conformation that spans the ~70 Å between the decoding center and the peptidyl transferase center. If the compact conformation observed crystallographically is physiologically relevant, this implies that either: 1) the extended and compact conformations are in equilibrium in solution and the ribosome captures the extended form; or 2) the compact conformation binds and then extends on the ribosome. To distinguish between these possibilities, Fu et al. used a rapid mixing technique paired with cryoEM to study the structural progression after mixing a UAA-programmed release complex (RC0) with RF-1. At 24 ms, 25% of ribosome-bound RF was in its compact form, in a sort of pre-accommodation state previously observed for RF-2 by cryo-EM in ribosomes stalled at the terminus of an mRNA lacking a stop codon131. This population rapidly decayed by 60 ms, into a ribosome-bound extended conformation RF complex (Fig. 5c), indicating 1) that RF-1 can bind in a compact state; and 2) that the conformation shift to the elongated state happens very rapidly upon RF binding, whereas the hydrolysis step, which acts to release the nascent polypeptide chain, occurs on a much longer time scale. The authors hypothesize that the compact form of RF may assist with rapid factor binding and more accurate stop-codon decoding.
Responding to stress: a glimpse at sequestration.
Nutrient starvation and other stress can lead to translational suppression, which directs cellular resources away from the energy-intensive process of protein production and thereby promotes cell survival132. In E. coli, hibernation factors work in concert to suppress protein synthesis by binding to and blocking functional sites on the SSU, and by inducing the reversible dimerization of 70S ribosomes into a translationally-inactive 100S sequestration particle133,134. In contrast to ribosome degradation, which has been observed in mammalian cells135, the generation of such a particle effectively allows cells to downregulate translation during starvation, and to then rapidly reinitiate translation should nutrients become available. The first structures of this sequestration particle were generated through cryoEM, and the visualization of these particles dimerized head-to-head through the SSU provided a structural understanding of translational repression in bacteria136–138. Interestingly, although hibernation factors vary between species, the process of sequestration may be conserved as a 110S sequestration particle has been identified in mammalian cells139.
What lies ahead.
Whereas single particle analysis of purified ribosomes has provided key mechanistic insights into ribosome biology, there are a number of benefits to studying samples in situ – that is, in their native cellular environment. By avoiding cell lysis and downstream purification, one can interrogate minimally perturbed ribosomes that are likely to better retain transiently bound cofactors. Moreover, in situ imaging can reveal sub-cellular localization patterns of sub-nanometer scale features that may be associated with biological function.
Studying ribosomes in situ via electron microscopy is typically accomplished using cryo-electron tomography (cryoET), often preceded by focused ion beam (FIB)-milling, which is used to thin a cellular sample to allow transmission of elastically scattered electrons. In contrast to cryoEM, cryoET repeatedly images the same ~50–300 nm thick sample at different stage tilt angles, which helps to disentangle particles at different depths along any one projection axis. Such a “tilt series” can then be back-projected to reconstruct a 3D tomogram of the field of view typically 0.05–0.5 μm3, thereby visualizing a significant portion of small bacterial cells, and a relatively small subregion of typical eukaryotic cells. From this low signal-to-noise ratio tomogram, ribosomes can be identified, extracted, aligned, and averaged to increase the signal-to-noise ratio and structural resolution in a process called sub-tomogram averaging (STA)140.
Until recently, cryoET and STA structures rarely exceeded ~1–2 nm resolution, which limited the field’s ability to glean molecular insights from the resulting structures. However, recent advancements in hardware for FIB-milling and imaging141,142, strategies for data collection143–145, and image processing software28,146–153 have poised structural biology for an “in situ resolution revolution” akin to the single particle analysis “resolution revolution” of the early 2010s7. Downstream tools to analyze structural heterogeneity have also expanded, allowing identification of rare and dynamic subpopulations of ribosomes in situ30,151,154–157. Taken together, the field now enjoys faster and easier data collections, larger and higher quality datasets, more powerful data processing, and novel data analysis opportunities.
While many of these developments are recent and their synergistic effects are likely to bear fruit over the next several years, several exciting findings have already hinted at what is to come (Fig. 6). Tegunov et al. demonstrated that STA could resolve chloramphenicol-treated M. pneumonaie ribosomes at 3.5Å resolution in situ28, and Xue et al. further analyzed the same system to characterize the distribution of translational states under antibiotic and untreated conditions in situ, resolving 13 compositionally and conformationally distinct states that recapitulate the known translation cycle118. Notably, detailed analysis of native polysomes in situ allowed Xue et al. to directly observe protein bL9 playing a role in polysome coordination and collision avoidance. In a novel approach to in situ imaging and processing, Lucas et al. designed 2D template matching (2DTM) which searches individual 2D micrographs for instances of a high-resolution template29. In one application of 2DTM they used a M. pneumonaie 50S subunit structure as a “bait”, identifying via local searches the variable presence of adjacent smaller 30S subunits. In principle this approach could enable quantitation of small subpopulations of ribosomes with a particular cofactor of interest bound. Finally, we have developed a deep-learning based approach to study structural heterogeneity among subtomograms and applied it to the M. pneumonaie dataset first reported by Tegunov et al. In addition to recapitulating many of the aforementioned insights, this tool additionally resolved a minor population (~2%) of ribosomes associated with the membrane, thereby simultaneously visualizing a moderate resolution structure of a 70S ribosome - SecYEG holotranslocon co-complex in situ30.
Figure 6. Cryogenic electron tomography (cryoET) enables new insights into translation in situ.
(a) Illustrative example of distinct ribosome translational states identified by cryoET sub-tomogram averaging and classification, mapped back to the cell and colored by translation state. Inset: Example of near-atomic resolution maps (gray, semi-transparent) from cryoET in situ, with fit atomic model pdb:7PHB (ribbon multi-color). Data were reprocessed from Xue et al., 2022118. (b) Distribution of translation cycle states calculated from classification results of three recent in situ studies, highlighting similarities in relative abundance of states associated with various stages of the translation cycle observed by Xue et al. 2022118 [1], Hoffman et al. 2022159 [2], and Gemmer et al. 2023160 [3].
With this rich toolkit to pursue structural studies in vitro and in situ, we anticipate a wave of new biologically insightful findings in the coming years. For example, one can now directly develop and test structure-function hypotheses in the native cellular environment, and subsequently elucidate the specific structural mechanism involved with ~10−2 second time resolution in vitro, all at near-atomic resolution. Such tools will be critical to address key remaining questions in the field, including: do ribosome biogenesis, translation, and sequestration, along with their associated cofactors, exhibit specific subcellular distributions; are such distributions perturbed in diseased states; and do the structural states captured in vitro accurately reflect states and their relative populations in the cell? We also posit that in situ approaches are likely to enable discovery of novel or weakly interacting cofactors not captured with traditional cryoEM using purified samples. We eagerly await the insights that lie ahead for the study of ribosome dynamics at the intersection of structural, molecular, and cellular biology enabled by traditional cryoEM, time-resolved cryoEM, and cryoET.
Acknowledgments.
This work was supported by NIH grants R01-GM144542, 5T32-GM007287, and NSF-CAREER grant 2046778. Research in the Davis lab is supported by the Whitehead Family and the Sloan Foundation.
References.
- 1.Cate J.H., Yusupov M.M., Yusupova G.Z., Earnest T.N., and Noller H.F. (1999). X-ray crystal structures of 70S ribosome functional complexes. Science 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., and Yusupov M. (2011). The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529. 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 3.Davis J.H., and Williamson J.R. (2017). Structure and dynamics of bacterial ribosome biogenesis. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 372. 10.1098/rstb.2016.0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shajani Z., Sykes M.T., and Williamson J.R. (2011). Assembly of bacterial ribosomes. Annu Rev Biochem 80, 501–526. 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 5.Wilson D.N., and Nierhaus K.H. (2007). The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol 42, 187–219. 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 6.Kinman L.F., Powell B.M., Zhong E.D., Berger B., and Davis J.H. (2023). Uncovering structural ensembles from single-particle cryo-EM data using cryoDRGN. Nat Protoc 18, 319–339. 10.1038/s41596-022-00763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlbrandt W. (2014). Biochemistry. The resolution revolution. Science 343, 1443–1444. 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 8.Faruqi A.R., and Henderson R. (2007). Electronic detectors for electron microscopy. Curr Opin Struct Biol 17, 549–555. 10.1016/j.sbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Scheres S.H. (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Punjani A., Rubinstein J.L., Fleet D.J., and Brubaker M.A. (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 11.Zhong E.D., Bepler T., Berger B., and Davis J.H. (2021). CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods 18, 176–185. 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J., Kinman L.F., Jahagirdar D., Ortega J., and Davis J.H. (2023). KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion. Nat Struct Mol Biol 10.1038/s41594-023-01078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palade G.E. (1955). A small particulate component of the cytoplasm. J Biophys Biochem Cytol 1, 59–68. 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells W.A. (2005). Ribosomes, or the particles of Palade. J. Cell Biol. 10.1083/jcb1681fta3. [DOI] [Google Scholar]
- 15.Luria S.E., Delbruck M., and Anderson T.F. (1943). Electron Microscope Studies of Bacterial Viruses. J Bacteriol 46, 57–77. 10.1128/jb.46.1.57-77.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller O.L. Jr., Hamkalo B.A., and Thomas C.A. Jr. (1970). Visualization of bacterial genes in action. Science 169, 392–395. 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- 17.Gotta S.L., Miller O.L. Jr., and French S.L. (1991). rRNA transcription rate in Escherichia coli. J Bacteriol 173, 6647–6649. 10.1128/jb.173.20.6647-6649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Narvaez C.C., and Schaup H.W. (1979). In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J Mol Biol 134, 1–22. 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 19.Brown A., and Shao S. (2018). Ribosomes and cryo-EM: a duet. Curr Opin Struct Biol 52, 1–7. 10.1016/j.sbi.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Lake J.A. (1976). Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J Mol Biol 105, 131–139. 10.1016/0022-2836(76)90200-x. [DOI] [PubMed] [Google Scholar]
- 21.Frank J., Verschoor A., and Boublik M. (1982). Multivariate statistical analysis of ribosome electron micrographs. L and R lateral views of the 40 S subunit from HeLa cells. J Mol Biol 161, 107–133. [DOI] [PubMed] [Google Scholar]
- 22.Verschoor A., Frank J., Radermacher M., Wagenknecht T., and Boublik M. (1984). Three-dimensional reconstruction of the 30 S ribosomal subunit from randomly oriented particles. J Mol Biol 178, 677–698. [DOI] [PubMed] [Google Scholar]
- 23.Frank J., Bretaudiere J.P., Carazo J.M., Verschoor A., and Wagenknecht T. (1988). Classification of images of biomolecular assemblies: a study of ribosomes and ribosomal subunits of Escherichia coli. J Microsc 150 ( Pt 2), 99–115. [DOI] [PubMed] [Google Scholar]
- 24.Carazo J.M., and Frank J. (1988). Three-dimensional matching of macromolecular structures obtained from electron microscopy: an application to the 70S and 50S E. coli ribosomal particles. Ultramicroscopy 25, 13–22. [DOI] [PubMed] [Google Scholar]
- 25.Radermacher M., Wagenknecht T., Verschoor A., and Frank J. (1987). Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 146 (Pt 2), 113–136. [DOI] [PubMed] [Google Scholar]
- 26.Gao H., Valle M., Ehrenberg M., and Frank J. (2004). Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol 147, 283–290. [DOI] [PubMed] [Google Scholar]
- 27.O’Reilly F.J., Xue L., Graziadei A., Sinn L., Lenz S., Tegunov D., Blotz C., Singh N., Hagen W.J.H., Cramer P., et al. (2020). In-cell architecture of an actively transcribing-translating expressome. Science 369, 554–557. 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tegunov D., Xue L., Dienemann C., Cramer P., and Mahamid J. (2021). Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 A in cells. Nat Methods 18, 186–193. 10.1038/s41592-020-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucas B.A., Himes B.A., Xue L., Grant T., Mahamid J., and Grigorieff N. (2021). Locating macromolecular assemblies in cells by 2D template matching with cisTEM. eLife 10. 10.7554/eLife.68946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell B.M., and Davis J.H. (2023). Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN. bioRxiv, 2023.2005.2031.542975. 10.1101/2023.05.31.542975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis J.H., Tan Y.Z., Carragher B., Potter C.S., Lyumkis D., and Williamson J.R. (2016). Modular Assembly of the Bacterial Large Ribosomal Subunit. Cell 167, 1610–1622 e1615. 10.1016/j.cell.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabuck-Gibbons J.N., Lyumkis D., and Williamson J.R. (2022). Quantitative mining of compositional heterogeneity in cryo-EM datasets of ribosome assembly intermediates. Structure 30, 498–509 e494. 10.1016/j.str.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M., and Ludtke S.J. (2021). Deep learning-based mixed-dimensional Gaussian mixture model for characterizing variability in cryo-EM. Nat Methods 18, 930–936. 10.1038/s41592-021-01220-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A., and Fleet D.J. (2021). 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J Struct Biol 213, 107702. 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 35.Jobe A., Liu Z., Gutierrez-Vargas C., and Frank J. (2019). New Insights into Ribosome Structure and Function. Cold Spring Harb Perspect Biol 11. 10.1101/cshperspect.a032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klinge S., and Woolford J.L. Jr. (2019). Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20, 116–131. 10.1038/s41580-018-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodson S.A. (2011). RNA folding pathways and the self-assembly of ribosomes. Accounts of chemical research 44, 1312–1319. 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaczanowska M., and Ryden-Aulin M. (2007). Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71, 477–494. 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duss O., Stepanyuk G.A., Puglisi J.D., and Williamson J.R. (2019). Transient Protein-RNA Interactions Guide Nascent Ribosomal RNA Folding. Cell 179, 1357–1369 e1316. 10.1016/j.cell.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traub P., and Nomura M. (1968). Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci U S A 59, 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traub P., Hosokawa K., Craven G.R., and Nomura M. (1967). Structure and function of E. coli ribosomes, IV. Isolation and characterization of functionally active ribosomal proteins. Proc Natl Acad Sci U S A 58, 2430–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima S., and Nomura M. (1970). Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226, 1214. [DOI] [PubMed] [Google Scholar]
- 43.Held W.A., Ballou B., Mizushima S., and Nomura M. (1974). Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem 249, 3103–3111. [PubMed] [Google Scholar]
- 44.Herold M., and Nierhaus K.H. (1987). Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J Biol Chem 262, 8826–8833. [PubMed] [Google Scholar]
- 45.Rohl R., and Nierhaus K.H. (1982). Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci U S A 79, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dohme F., and Nierhaus K.H. (1976). Total reconstitution and assembly of 50 S subunits from Escherichia coli Ribosomes in vitro. J Mol Biol 107, 585–599. [DOI] [PubMed] [Google Scholar]
- 47.Nierhaus K.H., and Dohme F. (1974). Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A 71, 4713–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nierhaus K.H. (1991). The assembly of prokaryotic ribosomes. Biochimie 73, 739–755. 0300–9084(91)90054–5 [pii]. [DOI] [PubMed] [Google Scholar]
- 49.Sieber G., and Nierhaus K.H. (1978). Kinetic and thermodynamic parameters of the assembly in vitro of the large subunit from Escherichia coli ribosomes. Biochemistry 17, 3505–3511. 10.1021/bi00610a013. [DOI] [PubMed] [Google Scholar]
- 50.Bunner A.E., Nord S., Wikstrom P.M., and Williamson J.R. (2010). The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J Mol Biol 398, 1–7. 10.1016/j.jmb.2010.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stokes J.M., Davis J.H., Mangat C.S., Williamson J.R., and Brown E.D. (2014). Discovery of a small molecule that inhibits bacterial ribosome biogenesis. eLife 3, e03574. 10.7554/eLife.03574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seffouh A., Trahan C., Wasi T., Jain N., Basu K., Britton R.A., Oeffinger M., and Ortega J. (2022). RbgA ensures the correct timing in the maturation of the 50S subunits functional sites. Nucleic Acids Res. 10.1093/nar/gkac059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jomaa A., Jain N., Davis J.H., Williamson J.R., Britton R.A., and Ortega J. (2014). Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res 42, 3419–3435. 10.1093/nar/gkt1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gulati M., Jain N., Davis J.H., Williamson J.R., and Britton R.A. (2014). Functional interaction between ribosomal protein L6 and RbgA during ribosome assembly. PLoS genetics 10, e1004694. 10.1371/journal.pgen.1004694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sashital D.G., Greeman C.A., Lyumkis D., Potter C.S., Carragher B., and Williamson J.R. (2014). A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli. eLife 3. 10.7554/eLife.04491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maksimova E., Kravchenko O., Korepanov A., and Stolboushkina E. (2022). Protein Assistants of Small Ribosomal Subunit Biogenesis in Bacteria. Microorganisms 10. 10.3390/microorganisms10040747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S.S., Sperling E., Silverman J.M., Davis J.H., and Williamson J.R. (2012). Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol Biosyst 8, 3325–3334. 10.1039/c2mb25310k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindahl L. (1975). Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol 92, 15–37. 0022–2836(75)90089–3 [pii]. [DOI] [PubMed] [Google Scholar]
- 59.Guo Q., Goto S., Chen Y., Feng B., Xu Y., Muto A., Himeno H., Deng H., Lei J., and Gao N. (2013). Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res 41, 2609–2620. 10.1093/nar/gks1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li N., Chen Y., Guo Q., Zhang Y., Yuan Y., Ma C., Deng H., Lei J., and Gao N. (2013). Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res 41, 7073–7083. 10.1093/nar/gkt423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ni X., Davis J.H., Jain N., Razi A., Benlekbir S., McArthur A.G., Rubinstein J.L., Britton R.A., Williamson J.R., and Ortega J. (2016). YphC and YsxC GTPases assist the maturation of the central protuberance, GTPase associated region and functional core of the 50S ribosomal subunit. Nucleic Acids Res 44, 8442–8455. 10.1093/nar/gkw678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabuck-Gibbons J.N., Popova A.M., Greene E.M., Cervantes C.F., Lyumkis D., and Williamson J.R. (2020). SrmB Rescues Trapped Ribosome Assembly Intermediates. J Mol Biol 432, 978–990. 10.1016/j.jmb.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nikolay R., Hilal T., Qin B., Mielke T., Burger J., Loerke J., Textoris-Taube K., Nierhaus K.H., and Spahn C.M.T. (2018). Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution. Mol Cell 70, 881–893 e883. 10.1016/j.molcel.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Qin B., Lauer S.M., Balke A., Vieira-Vieira C.H., Burger J., Mielke T., Selbach M., Scheerer P., Spahn C.M.T., and Nikolay R. (2023). Cryo-EM captures early ribosome assembly in action. Nat Commun 14, 898. 10.1038/s41467-023-36607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong X., Doerfel L.K., Sheng K., Rabuck-Gibbons J.N., Popova A.M., Lyumkis D., and Williamson J.R. (2023). Near-physiological in vitro assembly of 50S ribosomes involves parallel pathways. Nucleic Acids Res 51, 2862–2876. 10.1093/nar/gkad082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nikolay R., Hilal T., Schmidt S., Qin B., Schwefel D., Vieira-Vieira C.H., Mielke T., Burger J., Loerke J., Amikura K., et al. (2021). Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization. Mol Cell 81, 1200–1215 e1209. 10.1016/j.molcel.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 67.Chen S.S., and Williamson J.R. (2013). Characterization of the Ribosome Biogenesis Landscape in E. coli Using Quantitative Mass Spectrometry. J Mol Biol 425, 767–779. 10.1016/j.jmb.2012.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gamalinda M., and Woolford J.L. Jr. (2015). Paradigms of ribosome synthesis: Lessons learned from ribosomal proteins. Translation (Austin) 3, e975018. 10.4161/21690731.2014.975018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gor K., and Duss O. (2023). Emerging Quantitative Biochemical, Structural, and Biophysical Methods for Studying Ribosome and Protein-RNA Complex Assembly. Biomolecules 13. 10.3390/biom13050866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheng K., Li N., Rabuck-Gibbons J.N., Dong X., Lyumkis D., and Williamson J.R. (2022). Unsupervised Voxel-based Segmentation reveals a Landscape of Bacterial Ribosome Large Subunit Early Assembly. bioRxiv, 2022.2011.2009.515851. 10.1101/2022.11.09.515851. [DOI] [Google Scholar]
- 71.Jomaa A., Stewart G., Martin-Benito J., Zielke R., Campbell T.L., Maddock J.R., Brown E.D., and Ortega J. (2011). Understanding ribosome assembly: the structure of in vivo assembled immature 30S subunits revealed by cryo-electron microscopy. RNA 17, 697–709. 10.1261/rna.2509811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Z., Guo Q., Goto S., Chen Y., Li N., Yan K., Zhang Y., Muto A., Deng H., Himeno H., et al. (2014). Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence. Protein & cell 5, 394–407. 10.1007/s13238-014-0044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leong V., Kent M., Jomaa A., and Ortega J. (2013). Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 19, 789–802. 10.1261/rna.037523.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jomaa A., Stewart G., Mears J.A., Kireeva I., Brown E.D., and Ortega J. (2011). Cryo-electron microscopy structure of the 30S subunit in complex with the YjeQ biogenesis factor. RNA 17, 2026–2038. 10.1261/rna.2922311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo Q., Yuan Y., Xu Y., Feng B., Liu L., Chen K., Sun M., Yang Z., Lei J., and Gao N. (2011). Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A 108, 13100–13105. 10.1073/pnas.1104645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lopez-Alonso J.P., Kaminishi T., Kikuchi T., Hirata Y., Iturrioz I., Dhimole N., Schedlbauer A., Hase Y., Goto S., Kurita D., et al. (2017). RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket. Nucleic Acids Res 45, 6945–6959. 10.1093/nar/gkx324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Datta P.P., Wilson D.N., Kawazoe M., Swami N.K., Kaminishi T., Sharma M.R., Booth T.M., Takemoto C., Fucini P., Yokoyama S., and Agrawal R.K. (2007). Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell 28, 434–445. S1097–2765(07)00596–5 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boehringer D., O’Farrell H.C., Rife J.P., and Ban N. (2012). Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem 287, 10453–10459. 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stephan N.C., Ries A.B., Boehringer D., and Ban N. (2021). Structural basis of successive adenosine modifications by the conserved ribosomal methyltransferase KsgA. Nucleic Acids Res 49, 6389–6398. 10.1093/nar/gkab430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schedlbauer A., Iturrioz I., Ochoa-Lizarralde B., Diercks T., Lopez-Alonso J.P., Lavin J.L., Kaminishi T., Capuni R., Dhimole N., de Astigarraga E., et al. (2021). A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit. Sci Adv 7. 10.1126/sciadv.abf7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma M.R., Barat C., Wilson D.N., Booth T.M., Kawazoe M., Hori-Takemoto C., Shirouzu M., Yokoyama S., Fucini P., and Agrawal R.K. (2005). Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell 18, 319–329. S1097–2765(05)01221–9 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 82.Razi A., Davis J.H., Hao Y., Jahagirdar D., Thurlow B., Basu K., Jain N., Gomez-Blanco J., Britton R.A., Vargas J., et al. (2019). Role of Era in assembly and homeostasis of the ribosomal small subunit. Nucleic Acids Res 47, 8301–8317. 10.1093/nar/gkz571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seffouh A., Jain N., Jahagirdar D., Basu K., Razi A., Ni X., Guarne A., Britton R.A., and Ortega J. (2019). Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate. Nucleic Acids Res 47, 10414–10425. 10.1093/nar/gkz770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X., Yan K., Zhang Y., Li N., Ma C., Li Z., Zhang Y., Feng B., Liu J., Sun Y., et al. (2014). Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Res 42, 13430–13439. 10.1093/nar/gku1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang W., Li W., Ge X., Yan K., Mandava C.S., Sanyal S., and Gao N. (2020). Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation. Proc Natl Acad Sci U S A 117, 15609–15619. 10.1073/pnas.1914323117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng B., Mandava C.S., Guo Q., Wang J., Cao W., Li N., Zhang Y., Zhang Y., Wang Z., Wu J., et al. (2014). Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol 12, e1001866. 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Razi A., Britton R.A., and Ortega J. (2017). The impact of recent improvements in cryo-electron microscopy technology on the understanding of bacterial ribosome assembly. Nucleic Acids Res 45, 1027–1040. 10.1093/nar/gkw1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Connolly K., Rife J.P., and Culver G. (2008). Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol Microbiol 70, 1062–1075. 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inoue K., Basu S., and Inouye M. (2007). Dissection of 16S rRNA methyltransferase (KsgA) function in Escherichia coli. J Bacteriol 189, 8510–8518. 10.1128/JB.01259-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hopfield J.J. (1974). Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A 71, 4135–4139. 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frank J., and Gonzalez R.L. Jr. (2010). Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 79, 381–412. 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mitra K., and Frank J. (2006). Ribosome dynamics: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct 35, 299–317. [DOI] [PubMed] [Google Scholar]
- 93.Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., and Frank J. (2003). Locking and unlocking of ribosomal motions. Cell 114, 123–134. [DOI] [PubMed] [Google Scholar]
- 94.Gao H., Sengupta J., Valle M., Korostelev A., Eswar N., Stagg S.M., Van Roey P., Agrawal R.K., Harvey S.C., Sali A., et al. (2003). Study of the structural dynamics of the E coli 70S ribosome using real-space refinement. Cell 113, 789–801. [DOI] [PubMed] [Google Scholar]
- 95.Frank J., and Agrawal R.K. (2000). A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322. [DOI] [PubMed] [Google Scholar]
- 96.Mohan S., Donohue J.P., and Noller H.F. (2014). Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci U S A 111, 13325–13330. 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Valle M., Zavialov A., Li W., Stagg S.M., Sengupta J., Nielsen R.C., Nissen P., Harvey S.C., Ehrenberg M., and Frank J. (2003). Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10, 899–906. [DOI] [PubMed] [Google Scholar]
- 98.Seidelt B., Innis C.A., Wilson D.N., Gartmann M., Armache J.P., Villa E., Trabuco L.G., Becker T., Mielke T., Schulten K., et al. (2009). Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326, 1412–1415. 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C.L. 3rd, Ban N., and Frank J. (2005). Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324. 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ermolenko D.N., Majumdar Z.K., Hickerson R.P., Spiegel P.C., Clegg R.M., and Noller H.F. (2007). Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 370, 530–540. 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 101.Moazed D., and Noller H.F. (1989). Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148. 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 102.Munro J.B., Altman R.B., O’Connor N., and Blanchard S.C. (2007). Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell 25, 505–517. 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pan D., Kirillov S.V., and Cooperman B.S. (2007). Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 25, 519–529. 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Agrawal R.K., Penczek P., Grassucci R.A., and Frank J. (1998). Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Agrawal R.K., Penczek P., Grassucci R.A., Burkhardt N., Nierhaus K.H., and Frank J. (1999). Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem 274, 8723–8729. [DOI] [PubMed] [Google Scholar]
- 106.Allen G.S., Zavialov A., Gursky R., Ehrenberg M., and Frank J. (2005). The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121, 703–712. 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 107.Gao H., Zhou Z., Rawat U., Huang C., Bouakaz L., Wang C., Cheng Z., Liu Y., Zavialov A., Gursky R., et al. (2007). RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129, 929–941. 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 108.Frank J., Sengupta J., Gao H., Li W., Valle M., Zavialov A., and Ehrenberg M. (2005). The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579, 959–962. [DOI] [PubMed] [Google Scholar]
- 109.Bai X.C., McMullan G., and Scheres S.H. (2015). How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 40, 49–57. 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 110.Fischer N., Neumann P., Konevega A.L., Bock L.V., Ficner R., Rodnina M.V., and Stark H. (2015). Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature 520, 567–570. 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 111.Arenz S., Nguyen F., Beckmann R., and Wilson D.N. (2015). Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-A resolution. Proc Natl Acad Sci U S A 112, 5401–5406. 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Florin T., Maracci C., Graf M., Karki P., Klepacki D., Berninghausen O., Beckmann R., Vazquez-Laslop N., Wilson D.N., Rodnina M.V., and Mankin A.S. (2017). An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat Struct Mol Biol 24, 752–757. 10.1038/nsmb.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Paternoga H., Crowe-McAuliffe C., Bock L.V., Koller T.O., Morici M., Beckert B., Myasnikov A.G., Grubmuller H., Novacek J., and Wilson D.N. (2023). Structural conservation of antibiotic interaction with ribosomes. Nat Struct Mol Biol. 10.1038/s41594-023-01047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fromm S.A., O’Connor K.M., Purdy M., Bhatt P.R., Loughran G., Atkins J.F., Jomaa A., and Mattei S. (2023). The translating bacterial ribosome at 1.55 A resolution generated by cryo-EM imaging services. Nat Commun 14, 1095. 10.1038/s41467-023-36742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Herr A.J., Nelson C.C., Wills N.M., Gesteland R.F., and Atkins J.F. (2001). Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J Mol Biol 309, 1029–1048. 10.1006/jmbi.2001.4717. [DOI] [PubMed] [Google Scholar]
- 116.Smith A.M., Costello M.S., Kettring A.H., Wingo R.J., and Moore S.D. (2019). Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc Natl Acad Sci U S A 116, 21769–21779. 10.1073/pnas.1910613116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Atkins J.F., and Bjork G.R. (2009). A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev 73, 178–210. 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xue L., Lenz S., Zimmermann-Kogadeeva M., Tegunov D., Cramer P., Bork P., Rappsilber J., and Mahamid J. (2022). Visualizing translation dynamics at atomic detail inside a bacterial cell. Nature 610, 205–211. 10.1038/s41586-022-05255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cerullo F., Filbeck S., Patil P.R., Hung H.C., Xu H., Vornberger J., Hofer F.W., Schmitt J., Kramer G., Bukau B., et al. (2022). Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue. Nature 603, 509–514. 10.1038/s41586-022-04487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Saito K., Kratzat H., Campbell A., Buschauer R., Burroughs A.M., Berninghausen O., Aravind L., Green R., Beckmann R., and Buskirk A.R. (2022). Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria. Nature 603, 503–508. 10.1038/s41586-022-04416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., and Cate J.H. (2015). High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol 22, 336–341. 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dunkle J.A., and Cate J.H.D. (2011). The packing of ribosomes in crystals and polysomes. In Ribosomes: Structure, Function, and Dynamics, Rodnina M.V., Wintermeyer W., and Green R., eds. (Springer; Vienna: ), pp. 65–73. 10.1007/978-3-7091-0215-2_6. [DOI] [Google Scholar]
- 123.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., and Noller H.F. (2001). Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 124.Loveland A.B., Demo G., Grigorieff N., and Korostelev A.A. (2017). Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117. 10.1038/nature22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maeots M.E., and Enchev R.I. (2022). Structural dynamics: review of time-resolved cryo-EM. Acta Crystallogr D Struct Biol 78, 927–935. 10.1107/S2059798322006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carbone C.E., Loveland A.B., Gamper H.B. Jr., Hou Y.M., Demo G., and Korostelev A.A. (2021). Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP. Nat Commun 12, 7236. 10.1038/s41467-021-27415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dandey V.P., Budell W.C., Wei H., Bobe D., Maruthi K., Kopylov M., Eng E.T., Kahn P.A., Hinshaw J.E., Kundu N., et al. (2020). Time-resolved cryo-EM using Spotiton. Nat Methods 17, 897–900. 10.1038/s41592-020-0925-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kaledhonkar S., Fu Z., White H., and Frank J. (2018). Time-Resolved Cryo-electron Microscopy Using a Microfluidic Chip. Methods Mol Biol 1764, 59–71. 10.1007/978-1-4939-7759-8_4. [DOI] [PubMed] [Google Scholar]
- 129.Kaledhonkar S., Fu Z., Caban K., Li W., Chen B., Sun M., Gonzalez R.L. Jr., and Frank J. (2019). Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature 570, 400–404. 10.1038/s41586-019-1249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fu Z., Indrisiunaite G., Kaledhonkar S., Shah B., Sun M., Chen B., Grassucci R.A., Ehrenberg M., and Frank J. (2019). The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Nat Commun 10, 2579. 10.1038/s41467-019-10608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.James N.R., Brown A., Gordiyenko Y., and Ramakrishnan V. (2016). Translational termination without a stop codon. Science 354, 1437–1440. 10.1126/science.aai9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ishihama A. (1999). Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 4, 135–143. 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 133.Yamagishi M., Matsushima H., Wada A., Sakagami M., Fujita N., and Ishihama A. (1993). Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control. EMBO J 12, 625–630. 10.1002/j.1460-2075.1993tb05695.x.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Polikanov Y.S., Blaha G.M., and Steitz T.A. (2012). How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336, 915–918. 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.An H., and Harper J.W. (2018). Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20, 135–143. 10.1038/s41556-017-0007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kato T., Yoshida H., Miyata T., Maki Y., Wada A., and Namba K. (2010). Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure 18, 719–724. 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 137.Ortiz J.O., Brandt F., Matias V.R., Sennels L., Rappsilber J., Scheres S.H., Eibauer M., Hartl F.U., and Baumeister W. (2010). Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol 190, 613–621. 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Matzov D., Aibara S., Basu A., Zimmerman E., Bashan A., Yap M.F., Amunts A., and Yonath A.E. (2017). The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat Commun 8, 723. 10.1038/s41467-017-00753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Krokowski D., Gaccioli F., Majumder M., Mullins M.R., Yuan C.L., Papadopoulou B., Merrick W.C., Komar A.A., Taylor D., and Hatzoglou M. (2011). Characterization of hibernating ribosomes in mammalian cells. Cell Cycle 10, 2691–2702. 10.4161/cc.10.16.16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Asano S., Engel B.D., and Baumeister W. (2016). In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J Mol Biol 428, 332–343. 10.1016/j.jmb.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 141.Obr M., Hagen W.J.H., Dick R.A., Yu L., Kotecha A., and Schur F.K.M. (2022). Exploring high-resolution cryo-ET and subtomogram averaging capabilities of contemporary DEDs. J Struct Biol 214, 107852. 10.1016/j.jsb.2022.107852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wagner F.R., Watanabe R., Schampers R., Singh D., Persoon H., Schaffer M., Fruhstorfer P., Plitzko J., and Villa E. (2020). Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat Protoc 15, 2041–2070. 10.1038/s41596-020-0320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hagen W.J.H., Wan W., and Briggs J.A.G. (2017). Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol 197, 191–198. 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eisenstein F., Yanagisawa H., Kashihara H., Kikkawa M., Tsukita S., and Danev R. (2023). Parallel cryo electron tomography on in situ lamellae. Nat Methods 20, 131–138. 10.1038/s41592-022-01690-1. [DOI] [PubMed] [Google Scholar]
- 145.Peck A., Carter S.D., Mai H., Chen S., Burt A., and Jensen G.J. (2022). Montage electron tomography of vitrified specimens. J Struct Biol 214, 107860. 10.1016/j.jsb.2022.107860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.de Teresa-Trueba I., Goetz S.K., Mattausch A., Stojanovska F., Zimmerli C.E., Toro-Nahuelpan M., Cheng D.W.C., Tollervey F., Pape C., Beck M., et al. (2023). Convolutional networks for supervised mining of molecular patterns within cellular context. Nat Methods 20, 284–294. 10.1038/s41592-022-01746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Moebel E., Martinez-Sanchez A., Lamm L., Righetto R.D., Wietrzynski W., Albert S., Lariviere D., Fourmentin E., Pfeffer S., Ortiz J., et al. (2021). Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nat Methods 18, 1386–1394. 10.1038/s41592-021-01275-4. [DOI] [PubMed] [Google Scholar]
- 148.Rice G., Wagner T., Stabrin M., Sitsel O., Prumbaum D., and Raunser S. (2023). TomoTwin: generalized 3D localization of macromolecules in cryo-electron tomograms with structural data mining. Nat Methods 20, 871–880. 10.1038/s41592-023-01878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tegunov D., and Cramer P. (2019). Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods 16, 1146–1152. 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Castano-Diez D., Kudryashev M., Arheit M., and Stahlberg H. (2012). Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol 178, 139–151. 10.1016/j.jsb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 151.Himes B.A., and Zhang P. (2018). emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nat Methods 15, 955–961. 10.1038/s41592-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zivanov J., Oton J., Ke Z., von Kugelgen A., Pyle E., Qu K., Morado D., Castano-Diez D., Zanetti G., Bharat T.A.M., et al. (2022). A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. eLife 11. 10.7554/eLife.83724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen M., Bell J.M., Shi X., Sun S.Y., Wang Z., and Ludtke S.J. (2019). A complete data processing workflow for cryo-ET and subtomogram averaging. Nat Methods 16, 1161–1168. 10.1038/s41592-019-0591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vuillemot R., Rouiller I., and Jonic S. (2023). MDTOMO method for continuous conformational variability analysis in cryo electron subtomograms based on molecular dynamics simulations. Sci Rep 13, 10596. 10.1038/s41598-023-37037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Harastani M., Eltsov M., Leforestier A., and Jonic S. (2022). TomoFlow: Analysis of Continuous Conformational Variability of Macromolecules in Cryogenic Subtomograms based on 3D Dense Optical Flow. J Mol Biol 434, 167381. 10.1016/j.jmb.2021.167381. [DOI] [PubMed] [Google Scholar]
- 156.Harastani M., Eltsov M., Leforestier A., and Jonic S. (2021). HEMNMA-3D: Cryo Electron Tomography Method Based on Normal Mode Analysis to Study Continuous Conformational Variability of Macromolecular Complexes. Front Mol Biosci 8, 663121. 10.3389/fmolb.2021.663121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Frank G.A., Bartesaghi A., Kuybeda O., Borgnia M.J., White T.A., Sapiro G., and Subramaniam S. (2012). Computational separation of conformational heterogeneity using cryo-electron tomography and 3D sub-volume averaging. J Struct Biol 178, 165–176. 10.1016/j.jsb.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Macgregor H. (2012). Oscar Miller (1925–2012)—the technological genius of chromosome science. Chromosome Research 20, 317–318. 10.1007/s10577-012-9278-z. [DOI] [Google Scholar]
- 159.Hoffmann P.C., Kreysing J.P., Khusainov I., Tuijtel M.W., Welsch S., and Beck M. (2022). Structures of the eukaryotic ribosome and its translational states in situ. Nat Commun 13, 7435. 10.1038/s41467-022-34997-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gemmer M., Chaillet M.L., van Loenhout J., Cuevas Arenas R., Vismpas D., Grollers-Mulderij M., Koh F.A., Albanese P., Scheltema R.A., Howes S.C., et al. (2023). Visualization of translation and protein biogenesis at the ER membrane. Nature 614, 160–167. 10.1038/s41586-022-05638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]