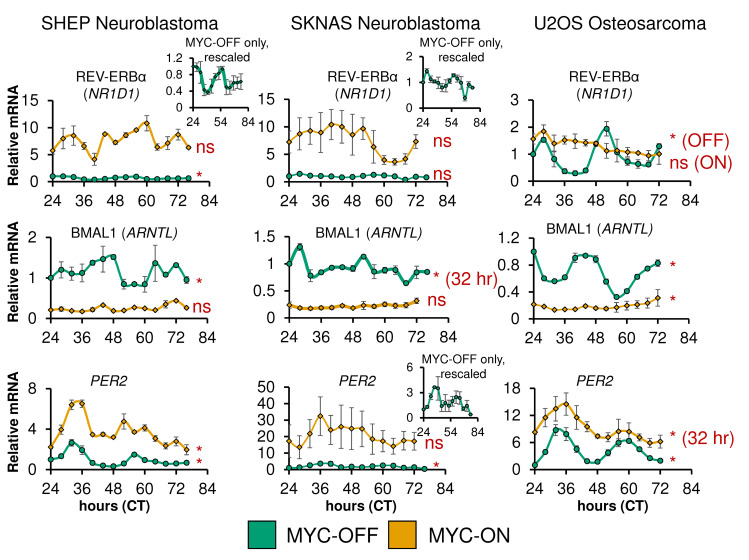
Fig 1. Oncogenic MYC disrupts molecular clock gene oscillation in cancer cells.
SHEP N-MYC-ER, SKNAS N-MYC-ER, and U2OS MYC-ER were treated with ethanol control (MYC-OFF) or 4-hydroxytamoxifen (MYC-ON) (4OHT) to activate MYC, and entrained with dexamethasone, and after 24 hours, RNA was collected every 2–4 hours for the indicated time period. Expression of the indicated genes was determined by quantitative PCR (qPCR), normalized to β2M. n = 2 biological replicates were performed, with error bars representing standard error of the mean (S.E.M.). CT = circadian time. Note the inset MYC-OFF only graphs for SHEP NR1D1, and SKNAS PER2, to show oscillation of these genes in MYC-OFF cells on a different scale. qPCR results were analyzed for rhythmicity by ECHO for both MYC-OFF and MYC-ON, with genes with a p value of < 0.05 and a BH.Adj.P.Value < 0.05 deemed rhythmic. For those with a period outside the 20–28 hour range, this is noted.