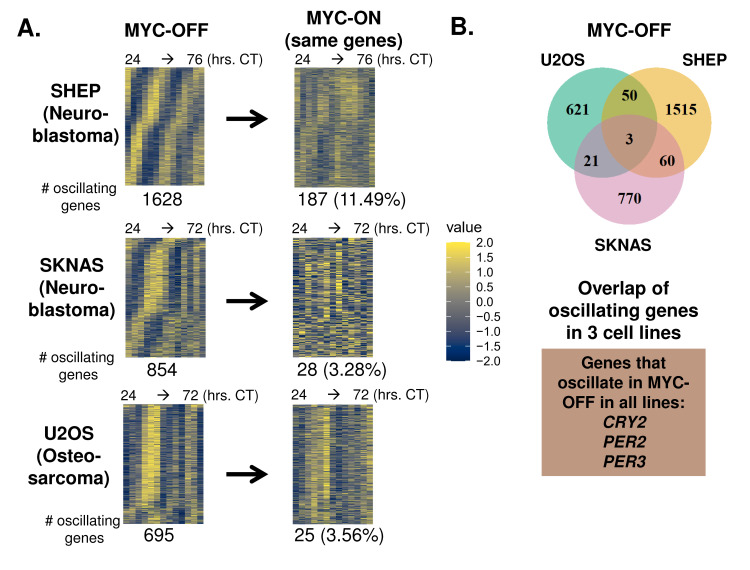
Fig 2. Oncogenic MYC disrupts global transcriptomic circadian oscillation in cancer cells.
A. RNA-sequencing was performed on the same n = 2 replicate SHEP N-MYC-ER, SKNAS-N-MYC-ER or U2OS MYC-ER time series ± 4OHT and + dexamethasone shown in Fig 1, with RNA samples collected every 2–4 hours at the indicated timepoints. RNA was analyzed for rhythmicity by ECHO for both MYC-OFF and MYC-ON, with genes with a 20–28 hour period and with p value < 0.05 and a BH.Adj.P.Value < 0.05 (SHEP, U2OS) or 0.1 (SKNAS) deemed rhythmic. These genes were sorted by phase and are presented in a heatmap for MYC-OFF. For MYC-ON, the same genes that are rhythmic in MYC-OFF are presented in the same order, but with MYC-ON values instead. B. The overlap of oscillating genes in MYC-OFF identified in (A) is shown by Venn diagram. Those genes that oscillated in all 3 cell lines are highlighted below the Venn diagram.