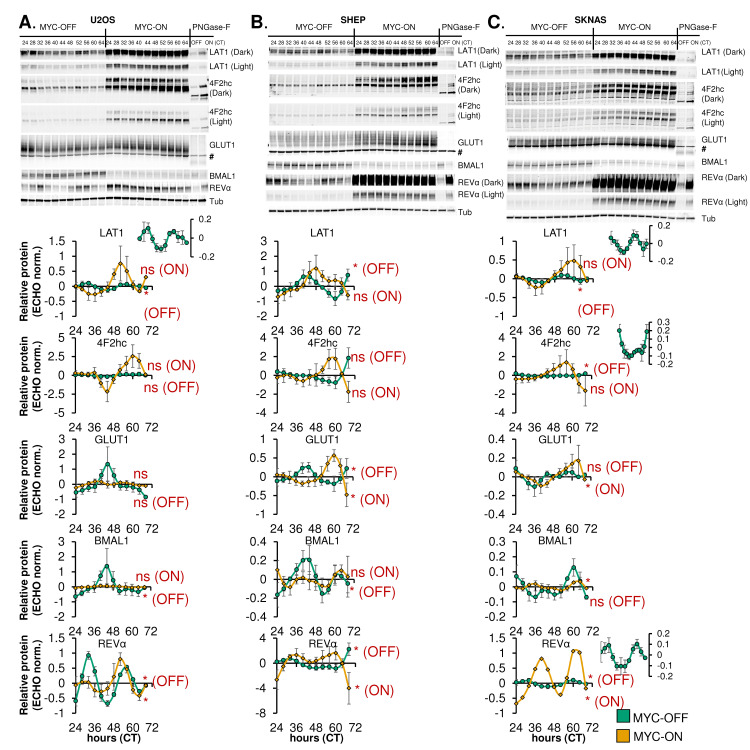
Fig 6. Rhythmic expression of nutrient transporters is disrupted by MYC.
A-C, top panels. U2OS MYC-ER (A), SHEP N-MYC-ER, (B), and SKNAS N-MYC-ER (C) were treated with ethanol control (MYC-OFF) or 4- hydroxytamoxifen (MYC-ON) (4OHT) to activate MYC, and entrained with dexamethasone, and after 24 hours, protein was collected every 4 hours for the indicated time period. Protein lysates were prepared to preserve protein glycosylation (see Methods), and immunoblot was performed for the indicated proteins. For some targets [4F2hc, LAT1, REV-ERBα (abbreviated REVα)], a darker exposure (‘dark’) and lighter exposure (‘light’) of the same blot are presented. For GLUT1, # indicates a non-specific band. Some samples (CT26 for U2OS, CT32 for SHEP and SKNAS) were treated with PNGase-F prior to immunoblot to remove glycosylation marks. Note that the PNGase-F lanes have less protein loaded than the other lanes. Data represent n = 3–4 biological replicates for each cell line. A-C, bottom panels. Results from n = 3–4 immunoblot replicates were quantified, relative to Tubulin, and analyzed in ECHO for circadian rhythmicity. Displayed curves are the baseline-subtracted and smoothed outputs from ECHO analysis. ECHO norm. = ECHO normalized. Proteins with a 22–26 hour period and a p value < 0.05 and a BH.Adj.P.Value < 0.05 were deemed rhythmic, which is indicated in the Figure. Note the inset MYC-OFF only graphs for U2OS LAT1, SKNAS LAT1, SKNAS 4F2hc, and SKNAS REV-ERBα, which show oscillation of these proteins in MYC-OFF cells on a different scale.