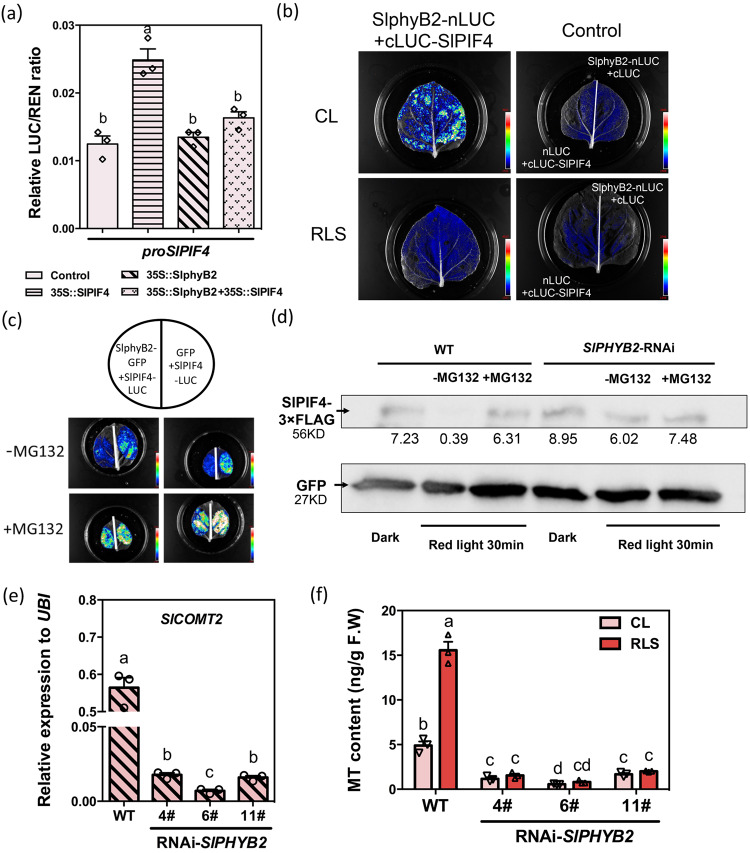
Fig. 5. Red light response of melatonin mediated by SlphyB2-SlPIF4-SlCOMT2.
a The transcriptional regulation relationship between SlphyB2 and SlPIF4. The dual-LUC experiment proves that SlphyB2 can inhibit the self-activation of SlPIF4 on its own promotor. Data are represented as Mean ± SEM (n = 3). b Quantitative analysis of luminescence intensity showing the interaction between SlphyB2 and SlPIF4 in Nicotiana benthamiana leaves. SlphyB2 interacts with the SlPIF4 protein, but the interaction disappears under red light. c SlphyB2 can ubiquitously degrade SlPIF4, and MG132 prevents the degradation. d Western blot detection of ubiquitination degradation of SlPIF4 mediated by SlphyB2. The addition of MG132 will inhibit the degradation of SlPIF4 by red light in WT, while in the interference strains of SlPHYB2, the bands of SlPIF4 are not different. GFP acts as an actin ensure consistent protein levels. This experiment was repeated independently two times with similar results. e Gene expression of SlCOMT2 in the SlPHYB2 interference lines. Samples were collected at Br+3. f Silence of SlPHYB2 makes the plant no longer be induced by the red light to produce more melatonin. The content of melatonin in wild tomato fruit was induced and accumulated by red light, but decreased in RNAi-SlPHYB2 lines, and was no longer induced by red light. Samples were collected at Br+3. Data in e, f are represented as Mean ± SEM (n = 3). In which 10–12 fruit collected from the same seedling were pooled as one biological replicate. For a, e, and f, the P values indicate the results from pairwise comparisons of one-way ANOVA tests. Different letters represent a significant difference at P < 0.05. Source data are provided as a Source Data file.