Abstract
Identified as the pathogenic genes of Alzheimer's disease (AD), APP, PSEN1, and PSEN2 mainly lead to early-onset AD, whose course is more aggressive, and atypical symptoms are more common than sporadic AD. Here, a novel missense mutation, APP E674Q (also named “Shanghai APP”), was detected in a Chinese index patient with typical late-onset AD (LOAD) who developed memory decline in his mid-70s. The results from neuroimaging were consistent with AD, where widespread amyloid β deposition was demonstrated in 18F-florbetapir Positron Emission Tomography (PET). APP E674Q is close to the β-secretase cleavage site and the well-studied Swedish APP mutation (KM670/671NL), which was predicted to be pathogenic in silico. Molecular dynamics simulation indicated that the E674Q mutation resulted in a rearrangement of the interaction mode between APP and BACE1 and that the E674Q mutation was more prone to cleavage by BACE1. The in vitro results suggested that the E674Q mutation was pathogenic by facilitating the BACE1-mediated processing of APP and the production of Aβ. Furthermore, we applied an adeno-associated virus (AAV)-mediated transfer of the human E674Q mutant APP gene to the hippocampi of two-month-old C57Bl/6 J mice. AAV-E674Q-injected mice exhibited impaired learning behavior and increased pathological burden in the brain, implying that the E674Q mutation had a pathogenicity that bore a comparison with the classical Swedish mutation. Collectively, we report a strong amyloidogenic effect of the E674Q substitution in AD. To our knowledge, E674Q is the only pathogenic mutation within the amyloid processing sequence causing LOAD.
Keywords: Alzheimer's disease, Amyloid beta, APP mutation, E674Q, Late onset
Introduction
Alzheimer's disease (AD [MIM: 104,300]) is the most common neurodegenerative disease and is characterized by amyloid plaques and neurofibrillary tangles, accounting for 60%–80% of dementia.1 AD can be classified as early-onset AD (EOAD) and late-onset AD (LOAD) based on the age at onset. Mutations in three genes (APP [MIM: 104,760], PSEN1 [MIM: 104,311], and PSEN2 [MIM: 600,759]), which encode amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2), respectively, usually result in EOAD. These cases are also called autosomal dominant Alzheimer's disease because of a dominant inheritance pattern, with more frequent noncognitive neurological manifestations.2,3 Mutation detection rates of APP, PSEN1, and PSEN2 are diverse across different ethnicities, in part because of different study recruitment criteria and selection bias.4, 5, 6, 7, 8, 9 Among them, mutations in PSEN1 are the most numerous, with a total number of more than 300 (https://www.alzforum.org). Furthermore, mutations in APP, PSEN1, and PSEN2 cause disease through different pathways. Mutations in APP are usually located within or adjacent to the amyloid β (Aβ) sequence10 and lead to overproduction of Aβ or accelerated Aβ aggregation. PSEN 1 and 2 are subunits of γ-secretase, and mutations in PSEN1 and PSEN2 often result in an increased ratio of Aβ42/Aβ40.11 In particular, the onset age of APP mutation carriers with an AD phenotype ranged from 40 to 60 in previous studies.10
Here, we report a novel missense mutation in APP (Shanghai APP), E674Q, in an index patient with late-onset familial AD. Molecular dynamics (MD) simulations determined the mutation's role in an APP-induced conformational switch of beta-site APP cleaving enzyme 1 (BACE1). We further investigated whether the E674Q mutation affects the cleavage of APP by BACE1 and the generation of Aβ in vitro and in vivo. In general, these findings provide proof of principle for the hypothesis that reducing BACE1-mediated APP fragmentation may protect against the disease, at least in those with APP mutations.
Materials and methods
Clinical information, neuropsychological examination, and neuroimaging
The patient visited the Memory Clinic of Ruijin Hospital affiliated to Shanghai Jiaotong University School of Medicine with a primary complaint of memory decline. Clinical examination, neuropsychological assessment, laboratory tests, and cranial magnetic resonance imaging (MRI) were performed as established procedures. Scales of neuropsychological assessment included Mini-Mental State Examination (MMSE), Montreal Cognitive Assessment (MoCA), and Addenbrooke Cognitive Examination-III (ACE-III). The diagnosis of AD was based on established criteria proposed by the National Institute on Aging-Alzheimer's Association workgroup (NIA-AA 2011).12 At later visits, 18F-fludeoxyglucose (FDG) and 18F-florbetapir positron emission computed tomography (PET) was also performed after intravenous injection of 18F-FDG (dosage: 6.0 mCi) and 18F-florbetapir (dosage: 5.91 mCi).
Genetic sequencing and mutation analysis
This study was approved by the ethics committee of Ruijin Hospital and informed consent was obtained. We conducted APP, PSEN1, and PSEN2 sequencing and apolipoprotein E (APOE) genotyping from DNA extracted from peripheral venous blood according to our previous research methods.7,13 Mutations were defined as rare variants with minor allele frequency < 0.01 according to the Genome Aggregation Database. A novel mutation was defined by having no record in the AlzForum Database (https://www.alzforum.org/mutations) or PubMed. The following online tools were used to predict deleterious mutations: SIFT (https://hpc.ilri.cgiar.org/sift-software); PROVEAN (http://provean.jcvi.org/index.php); PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2); Mutation Taster (http://www.mutationtaster.org); and CADD (https://cadd.gs.washington.edu/snv).
Computational modeling
In this study, an APP fragment with 30 residues (657–686) was considered. This fragment contained the known APP-cleavage site, which was centered in the fragment. APP is a classical type I multidomain protein with a single transmembrane span. The APP transmembrane domain contains the sequence of β-amyloid peptides.14 The in silico APP fragment was modeled as a helix. The structure of the APP peptide was energy minimized in Maestro 2020 using the OPLS_2005 force field. The APP fragment was minimized in water for 50 steps using the steepest descent method, followed by 200 steps using the conjugate gradient method. The mutant APP fragment was constructed based on the modeled wild-type APP fragment using Discovery Studio 2019. The subsequent optimization was consistent with the previous steps.
Protein-peptide docking operation
Protein-peptide docking was carried out using Maestro 2020 and Rosetta FlexPepDock. The structure (Protein Data Bank code: 2ZHT) of BACE1 at pH 4.5 was docked to the APP fragment. The BACE1 structure downloaded from the Protein Data Bank code database was visually checked and prepared by using the Protein Preparation Wizard encoded in the Maestro 2020, and pKa values were calculated with the PROPKA algorithm (http://propka.org) at pH = 4.5. We used Protein‒Protein Docking in Maestro 2020 to dock the APP fragment with BACE1. Parameters were set as defaults. High-scoring complexes were inspected visually to identify the most reasonable solution. The complex structure was further refined in Rosetta FlexPepDock.15,16
Molecular dynamics (MD) simulation
MD simulation was performed using the GROMACS 5.1.2 package, utilizing the OPLS/AA force field. Four systems, wild-type APP fragment-bound BACE1 (WT system), E674Q (Shanghai) mutant, KM670/671NL (Swedish) mutant, APP fragment-bound BACE1 (mutant system), and the free BACE1 (apo-BACE1) were solvated in a cubic SPC water box with at least 10 Å from the box boundary to any residue. Sodium and chloride ions were added to neutralize the four systems to an ionic concentration of 0.15 mol/L. All systems were minimized and gradually equilibrated in an NPT ensemble at 310 K and 1 bar. Periodic boundary conditions were applied to these simulation systems. Bonds connected to hydrogen atoms were restrained with the LINCS algorithm to allow an integration time step of 2 fs. Electrostatic interactions were calculated using the particle mesh Ewald method. Finally, the MD simulations were performed for 500 ns in each of the four systems. All analyses of MD trajectories were performed using tools implemented in the GROMACS 5.1.2 package.16 All the structural graphics were processed with PyMOL software.17
Cell culture and cell treatment
HEK293 cells were grown in Dulbecco's modified Eagle medium (Gibco) with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in 5% CO2. The cells were transiently transfected with plasmids encoding GFP-Vector, GFP-APPwt, GFP-APP E674Q, and GFP-APPswe using polyethylenimine (Sigma). The cells and medium were harvested 48 h after transfection. The expression of APP and BACE1-derived APP fragments was determined by Western blotting. Concentrations of Aβ were determined by enzyme-linked immunosorbent assay (ELISA).
Western blotting
Cells were lysed in RIPA buffer (Cat# P0013B, Beyotime) on ice for 40 min with occasional vortexing, and a protease inhibitor cocktail (Cat# P9599, Sigma) was added before use. Cell lysates were then centrifuged at 13,000 g for 15 min at 4 °C. The supernatant was collected, and protein concentration was quantified by BCA protein assay before boiling in the SDS loading buffer. The samples were separated by SDS-PAGE and then transferred to a nitrocellulose membrane. The membrane was blocked in 5% milk for 1 h at room temperature and incubated with primary antibodies including anti-GFP (Cat# sc-9996, Santa Cruz Biotechnology) and anti-Tubulin (Cat# MABT205, Millipore) overnight at 4 °C. The membranes were washed 3–5 times with TBST and labeled with horseradish peroxidase (HRP)-conjugated secondary antibodies. Signals were detected by enhanced chemiluminescence (ECL).
Aβ40 and Aβ42 ELISA
The concentrations of Aβ40 and Aβ42 in the cell culture medium were analyzed using human Aβ40 and Aβ42 ELISA kits (Cat# KHB3481 and Cat# KHB3544, Invitrogen) according to the manufacturer's instructions. Briefly, the conditioned medium was treated with a protease inhibitor cocktail and the serine protease inhibitor AEBSF. Then the medium was added to a 96-well plate coated with anti-Aβ 40 or anti-Aβ 42 antibody, together with the detection antibody, and incubated for 3 h. The plates were washed 4 times with washing buffer. The plate was incubated with anti-IgG-HRP and then washed according to the manufacturer's instructions. The signals were developed with stabilized chromogen. Recombinant human Aβ was used to develop the standard curve. The Aβ40 and Aβ42 concentrations were determined by comparison with the standard curve.
Transmission electron microscopy (TEM) micrograph of amyloid-beta 42 systems
Synthetic beta-amyloid peptide 42 carrying the E674Q mutation (DAQFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV) and the wild-type beta-amyloid peptide 42 (DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV) were purchased from QYAOBIO, China. The freshly prepared Aβ42 peptides of wild type and E674Q Aβ42 peptides were aggregated in HEPES buffer. Ten microliters of these solutions were added to carbon-coated, 100-mesh copper grids (Electron Microscopy Sciences, Hatfield, PA). The extra water was blotted with tissue paper after 2–3 min, and a few mild washings were performed with Milli-Q water (Millipore, Billerica, MA). A quantity of 0.1% of uranyl acetate was added for 5 min for sample staining. The grids were dried under an infrared lamp and examined with a LIBRA 120 EFTEM (Carl Zeiss, Oberkochen, Germany).17
Thioflavin-S staining characterizes Aβ42 aggregation
The freshly prepared Aβ42 peptides (20 μM) were added to a 200 μL sample containing 50 mM glycine and 25 μM thioflavin S. The samples were loaded in a 96-well plate and florescent intensity of each sample was measured by excitation at 440 nm and emission at 490 nm every 3 min at 37 °C for 12 h.17
AAV-injected animal model and stereotaxic injection
Animal experiments were performed according to the protocols and guidelines and were approved by the Ethics Committee of Shanghai Jiao Tong University School of Medicine. Adeno-associated virus vectors (AAVrh.9) encoding human mutant full-length APPswe, APPE674Q, APPwt, and empty control were purchased from Sangon Biotech, China. Eight-week-old C57BL/6 J male mice (GemPharmtech, China) were divided into 4 groups. After anesthetization with isoflurane, these vectors were injected bilaterally into the CA1 of the hippocampus of C57BL/6 J mice with the following coordinates: −2 mm anteroposterior, ±1 mm mediolateral, and −2 mm dorsoventral. Each site was injected with 2 μL (4 × 109 vg/site) of diluted AAV virus for 10 min.
Animal behavioral tests
A novel object recognition test was performed on the mice 6 months after AAV injection. The size of the open field is 55 × 55 × 30 cm with the bottom surface divided into 25 squares (11 × 11 cm). Each mouse was examined in an open field with two consecutive trials separated by a 24 h interval. The first trial (habituation phase) comprised an exploration of two identical objects (7 × 4 × 4 cm) placed in distant areas (noted as NEW area and OLD area), respectively, within the open field for 5 min. In the second trial (testing phase), the objects in NEW area were replaced by a new one with a different shape. The mouse was allowed to explore the changed pair of objects for 5 min. Trials 1 and 2 were recorded by SuperMaze, a video tracking system. The time spent in the OLD area (To) and the NEW area (Tn) in trial 1 and trial 2 were recorded, respectively. The discrimination ratio (DR) represented the ability of mice to explore new objects. DR was defined as the time spent in the OLD area (To) divided by the time spent in the NEW area (Tn). The DR in trial 1 and trial 2 were noted as DR1 and DR2, respectively.
Y-maze test was performed after the novel object recognition test. The Y-maze device is comprised of three arms (30 × 8 × 15 cm) with the included angle of 120°. In the computer software, the three arms are marked as 1, 2, and 3, respectively. Directly above, there is a camera that can cover the whole Y maze device to record the behavior track of the mice. The mice were placed on the fixed arm, and the mice were free to explore for 8 min. The total number and the order of the entered arms were recorded. The order of the first two entered arms was not calculated and started from the third time, the entered arm which was different from the first two was recorded as correct. The result was shown as an alternative behavior.
Barnes maze was performed on a circular platform with a diameter of 122 cm and strong light stimulation. There are 20 holes around the platform with a diameter of 5 cm. One of the holes (called the target hole) is associated with a dark box (the target box). Other holes are empty and not connected to any object. The dark box is set up as a drawer for easy escapement. The target box is not visible from the platform surface. The four quadrants of the platform are set with spatial cues of different shapes. The maze is elevated 140 cm. Animals can freely enter the target hole to escape. Before the test, all the mice were settled into the dark box for 1 min to adjust, and then mice were placed in the center of the circular platform and let them explore freely for 5 min. The habituation phase (days 1–5): In this phase, the overall spatial cue is fixed with respect to the target box every day, but the connection between the hole and the target box is not fixed. Each mouse was tested twice a day, the training time is 3 min each time. If the mouse finds the target hole within 3 min, it will stop recording and stop the strong light stimulation. If it cannot be found within 3 min, it will be guided by humans and kept in the target box for 60 s and then returned to the cage.
In the testing phase, the dark box was removed, and all mice were allowed to explore the platform freely for 3 min. SuperMaze system records the latency before entering the target hole. The whole process needs to be stimulated by strong light, and the untrained mice rest in a dark environment throughout the whole process.
Statistical analysis
GraphPad Prism 8 was applied for analysis of the results of in vitro and in vivo experiments. Analysis of variance (ANOVA) was used in comparisons across more than two groups. Tukey's multiple comparisons test was used in multiple comparisons between each group for post hoc analysis. Statistical significance was defined as P < 0.05. Data were presented as mean ± standard error of the mean (SEM).
Results
Patient information
The male subject of this report was 77 years old (16 years of education) when he first visited the Memory Clinic of Ruijin Hospital with memory decline over approximately three years. Amnestic symptoms had been apparent in recent months, such as forgetting to take medicine and repeating questions. One of his first-degree relatives had a similar medical history (Fig. 1A). His mother (Ⅰ-2) manifested cognitive symptoms after a stroke in her eighties and progressed to vascular dementia in her nineties. One of his siblings, a brother two years older than him (Ⅱ-3), progressed to Alzheimer's dementia and died in his late seventies. Laboratory tests showed no abnormalities, and other diseases were denied.
Figure 1.
Clinical details. (A) Family pedigree of the proband (II-4, black arrow) harboring APP E674Q. One of his brothers (Ⅱ-3) was diagnosed with Alzheimer's dementia. A younger brother (Ⅱ-6) passed away because of acute pancreatitis and complications. The subject's son (Ⅲ-1) was 45 years old and had good performance in a cognition examination (with 16 year-education, MMSE 30/30, MoCA 26/30). (B–E) Neuroimaging of the index patient. (B, C) Medial temporal lobe atrophy was evident in MRI at 33 months (MTA = 2 to MTA = 3). (D, E) PET 3D-reconstructed images show hypometabolism in bilateral frontal, parietal, and temporal lobes (D, 18F-FDG); widespread cerebral Aβ deposition (E, 18F-florbetapir).
The patient (II-4) at 77 years of age showed cognitive impairment based on scores of MMSE (23/30) and MoCA (18/30). The results of ACE-III (total: 66/100) indicated prominent memory impairment (memory domain: 9/26), along with a decline in time orientation (attention domain: 14/18) and language fluency (5/14). Linguistic (language domain: 23/26) and visuospatial (visuospatial domain: 15/16) functions were well maintained, as well as subitems such as calculative ability and naming. MRI showed atrophy of the bilateral frontotemporal lobes and the hippocampus, with multiple lacunar infarctions in the frontoparietal and basal ganglia. At last visit, this patient was bedridden with rapid cognitive decline. According to the medial temporal lobe atrophy (MTA) Scale,18 atrophy was graded with a score of 3 (Fig. 1C) due to obviously decreased hippocampal height compared with a score of 2 (MTA Scale score) 33 months prior (Fig. 1B). An 18F-FDG PET scan performed at 77 years of age (Fig. 1D) revealed asymmetric decreased metabolism. Another PET scan using 18F-florbetapir showed widespread amyloid deposition in the cortex of almost all cerebral lobes (Fig. 1E).
Genetic and molecular analysis
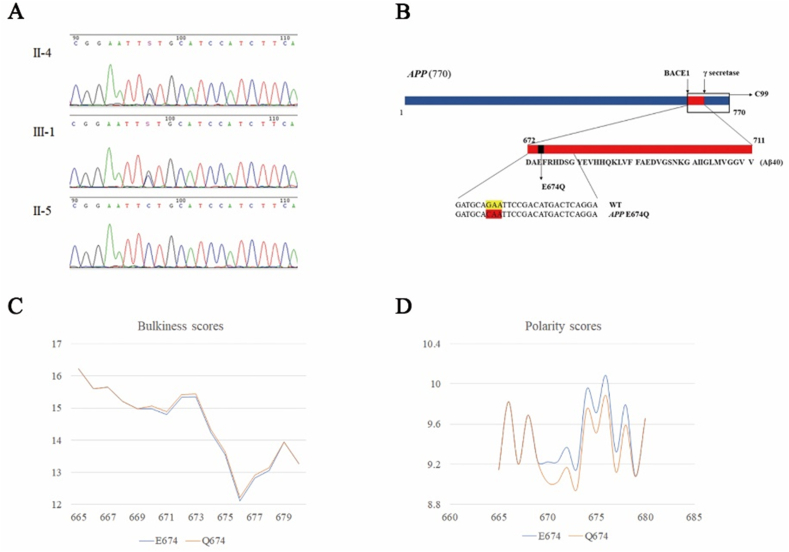
A novel APP mutation (NM_000484: exon16: c.2020G > C: p. E674Q) was identified in the index patient (Fig. 2A), with the APOE ε3/ε4 genotype. No other mutation was detected in PSEN1 and PSEN2. His son (Ⅲ-1), rather than his mother, also carried the APP E674Q mutation, as verified by Sanger sequencing (Fig. 3A). Base substitution (G replaced by C) resulted in a coding change of glutamic acid (Glu, E) to glutamine (Gln, Q) in codon 674. The frequency in the population of this variant is 0.003% (rs752361848) and is not recorded in the East Asian population. The mutation was not found in the AlzForum database, and no other mutation was reported at the same site. It was predicted to be damaging or probably damaging (or disease-causing) by SIFT, PolyPhen-2, Mutation Taster, and CADD (Phred-scaled score = 25.2). The PROVEAN tool indicated a neutral variant with a prediction score of −1.12 (cutoff = −2.5). In summary, four out of five prediction tools determined this mutation to be pathogenic. According to the algorithm proposed by Guerrerio and his colleagues,19 mutation E674Q can be classified as definite pathogenic.
Figure 2.
APP E674Q. (A) Sequencing results of APP E674Q. The index patient (II-4) and his son (III-1) harbored the mutation (NM_000484: exon16: c.2020G > C: p. E674Q), labeled with a black arrow. This mutation was negative in the subject's spouse (II-5). (B)APP gene and context of the amino acid substitution caused by the mutation. (C, D) Alterations in amino acid physical and chemical properties within the local primary sequence context.
Figure 3.
Conformational fluctuations of BACE1 in the MD simulations for apo, WT, KM670/671NL, and E674Q APP-BACE1 systems. The plot of PCA analysis for the BACE1 structure in the (A) apo system (B) WT system (C) E674Q system, and (D) KM670/671NL system. (E) RMSF of BACE1 in the four systems. (F) The BACE1 flap region in superimposed structures of the four systems. (G–J) The relative positions of residues ASP32, THR72, THR329, and ARG235 in the (G) apo system (H) WT system (I) E674Q system, and (J) KM670/671NL system. (K) Distance between ASP32 and THR72 tracked throughout the simulated time. (L) Distance between THR329 and THR72. (M) Distance between ARG235 and THR72. The apo BACE1 system is represented in grey, the WT system in blue, and the E674Q system in pink.
Of note, this mutation site (codon 674) is located just two amino acids away from the β-secretase cleavage site at the beginning of the Aβ sequence (672DAEFR …), indicating that E674Q might be a pathogenic gene mutation that affects β-secretase cleavage (Fig. 2B). The hydrophobicity score of the mutated peptide was unchanged with a minor alteration in bulkiness (Fig. 2C). Polarity was calculated to decrease from 9.944 to 9.744 in this residue by Expasy tools (Fig. 2D).
APP-induced conformational switch of BACE1 from MD simulations
MD simulations were carried out to explore the APP-induced conformational switch of BACE1. Four systems, apo-BACE1, wild type APP fragment-bound BACE1 (WT BACE1), KM670/671NL APP fragment-bound BACE1 (APP-Swedish mutant BACE1), and E674Q APP fragment-bound BACE1 (Shanghai mutant BACE1) were performed within the course of 500 ns simulation for each system, respectively. Gromos conformational cluster analysis was used to obtain the representative structures of the four systems within the course of a 500 ns simulation. The conformational changes of BACE1 were studied first. Root mean square fluctuation (RMSF) values were calculated for each of the four systems (Fig. 3E). Principal component analysis (PCA) was applied to identify the large-scale motions of BACE1 during the simulation. The first principal component was plotted (Fig. 3A–D). The regions insert A (Gly158-Leu167), insert C (Ala251-Pro258), insert D (Trp270-Thr274), insert E (Glu290-Ser295), and insert F (Asp311Asp317) in apo-BACE1 underwent large fluctuations, while those in the WT system and E674Q and KM670/671NL mutant systems were relatively rigid, which was consistent with the RMSF results. In terms of the WT system, the insert C and insert D regions of BACE1 moved toward the catalytic cavity, while insert F and insert A of BACE1 moved downward, which was suggested to enable the substrate to enter into the catalytic cavity. The 10 s loop (Lys9-Tyr14), located directly below the flap, was observed to flip down. In contrast, the conformational changes in insert C and insert D in the E674Q system and KM670/671NL system were similar to those in the apo-BACE1 system. The upward movement of insert-F and insert-A of BACE1 in the KM670/671NL mutant system (Fig. 3D) was different from that in other systems. The downward movement of insert F and insert A of BACE1 in the E674Q mutant system (Fig. 3C) was smaller than that in the WT system (Fig. 3B) by 2.6 Å. The 10 s loop remained unchanged in the mutant systems.
The opening and closing of the flap (Val67-Glu77) region were critical to the substrate binding and catalytic activity of BACE1 since the flap region is located just above the catalytic cavity of BACE1. The representative structures of the four systems were superimposed and the flap regions in each were highlighted (Fig. 3E). The BACE1 flap region was found to swing greatly in apo-BACE1, while it was less flexible upon the binding of the substrate in both the WT and E674Q mutant systems. However, the RMSF value representing the flap's range of motion in the E674Q system was higher than that in the WT system. Compared with apo-BACE1, the flap moved inward in the WT system by 5.1 Å and moved outward in the E674Q system by 3.4 Å.
Next, the four systems' conformational changes in the catalytic cavity were monitored. As shown in Figure 3K–M, distances between Cα(THR72) and Cβ (ASP32), between Cα(THR72) and Cα (THR329), and between OG1 (THR72) and NH1(ARG235) were used to monitor the change of the shape and size of the catalytic cavity. In the WT system, the three distances decreased compared with those in the apo-BACE1 system. In addition, an increase in each of the three distances was found in the E674Q and KM670/671NL systems. Regarding the KM670/671NL system, the distances between Cα(THR72) and Cβ(ASP32), between Cα(THR72) and Cα(THR329), and between OG1 (THR72) and NH1(ARG235) were 1.702 Å, 1.636 Å, and 1.892 Å, respectively. Regarding the E674Q system, the distances were 2.063 Å, 2.067, and 2.171 Å, respectively, which indicated a slight increase compared to the KM670/671NL system. The catalytic cavities of the four systems are displayed in Figure 3G–J.
The interaction mode of BACE1 with the APP fragment was then analyzed. As the simulation evolved, the E674Q APP fragment, similar to the KM670/671NL APP fragment, formed relatively fewer hydrogen bonds with BACE1 than it did in the WT system (Fig. 4A). The residue THR72, located at the tip of the flap, was identified to play an important role in the interaction with WT and mutant APP fragments. The hydroxyl group in THR72 formed a hydrogen bond with the amide group of the backbone in Asp672 (P1′) in the WT system (Fig. 4C), which was consistent with a previous study. In contrast, such an interaction was not found in the two mutant systems, in which the side chain of THR72 flipped up and the NH group of the backbone at THR72 formed a hydrogen bond with the oxygen of the carbonyl group in GLN674 (P3′) in the E674Q system and Ser679 (P4′) (Fig. 4C, D). Such a change in the interaction mode resulted in the eversion of the BACE1 flap in the mutant systems.
Figure 4.
The interaction between APP fragment and BACE1 in MD simulations. (A) Numbers of hydrogen bonds between BACE1 and the substrate (WT, E674Q, and KM670/671NL APP) monitored during the simulation. (B–D) Interaction between THR72 in BACE1 and the substrate (blue for the WT system, pink for the E674Q system, and purple for the KM670/671NL system). (E) The catalytic aspartate dyad in superimposed cluster structures of the four systems (the apo system and surrounding residues are represented in grey, the WT system and surrounding residues in blue, the E674Q system and surrounding residues in pink, and the KM670/671NL system and surrounding residues in green). (F) Proposed BACE1-mediated cleavage process of E674Q compared to WT APP. The WT system is represented as light blue. The E674Q mutant system is shown in purple. Aβ is represented in pink.
The conformational changes of the above key structural regions in the WT system versus the mutant systems resulted in a different state pertaining to the catalytic cavity of BACE1. This cavity was in the “closed state” in the WT system and the “open state” for BACE1 in the E674Q mutant system.
The representative structures of the WT and E674Q systems were used to calculate the binding free energy. X-score 1.3 was applied to compute the binding free energy.20 Finally, the interactions of the catalytic aspartate dyad in BACE1 with WT and mutant APP fragments were carefully examined. The catalytic aspartate dyads in each molecular system were superimposed. This result demonstrates that the catalytic aspartate dyad is close to the cleavage site of APP (M671-D672) in the E674Q mutant system, while it is far away in the WT system (Fig. 4E).
The E674Q mutation facilitates the fragmentation of APP by BACE1 in vitro
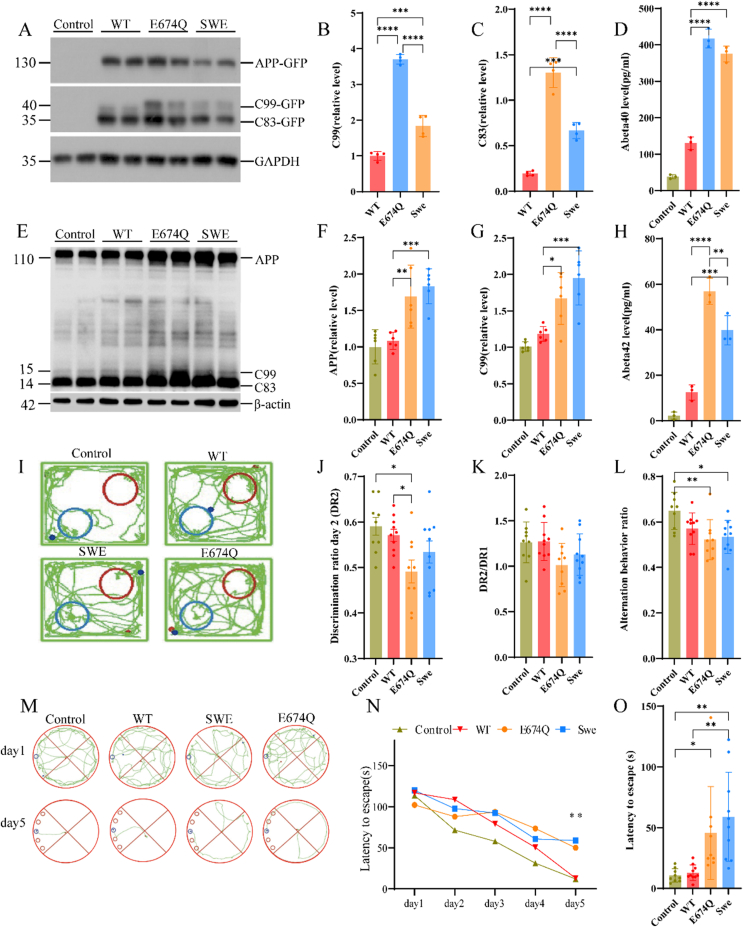
The cleavage of APP by BACE1 is the rate-limiting step in Aβ production.21 To investigate whether the E674Q mutation affects the cleavage of APP by BACE1 and the generation of Aβ, we transfected HEK293 cells with GFP-tagged wild-type APP, APP E674Q, and APPswe into HEK293 cells and determined the production of C99 and C83. Interestingly, we found that the production of C99, which results from BACE1-mediated cleavage, was increased by 2.38- and 1.95-fold in APP E674Q- and APPswe-transfected cells, respectively, compared with APP WT-transfected cells. The cells transfected with the APP E674Q mutation showed even higher C99 production than those transfected with the APPswe mutation (Fig. 5A, B). In line with these results, the media concentrations of Aβ40 and Aβ42 were increased in the APP E674Q and APPswe groups compared with the wild-type APP group, with the APP E674Q mutation showing higher Aβ40 and Aβ42 levels than the APPswe group (Fig. 5D, H). These results indicate that the E674Q mutation may promote AD by facilitating the BACE1-mediated processing of APP and the production of Aβ.
Figure 5.
The E674Q mutation facilitates the fragmentation of APP by BACE1 in vitro and in vivo. (A) Western blot analysis of HEK293 cells transfected with wild-type (WT) APP, APP E674Q, and APP with the Swedish mutation. (B, C) The levels of C99 and C83 fragments were increased in cells transfected with APP E674Q and APPswe. (D, H) ELISA quantification of the concentrations of Aβ40 (D) and Aβ42 (H) in conditioned medium from HEK293 cells transfected with wild-type and mutant APP. Data are presented as mean ± S.E.M. P < 0.01, one-way ANOVA. (E) Western blot analysis of AAV-mediated injection mice with APPwt, APPE674Q, and APPswe (Y188, ab32136, Abcam). (F, G) The levels of APP and C99 fragment were increased in mice hippocampus injected with AAV-APPE674Q and AAV-APPswe. (I–O) Behavioral experiments 6 months after the mice were injected. In the novel object recognition test, DR2 of APPE674Q animals was less than the control (P < 0.05; J, K); in the Y maze test, APPswe (P < 0.05) and APPE674Q (P < 0.01) had a significant difference from the control but no noticeable difference from APPwt (L); in the Barnes maze test, the two APP mutation groups show a potentially faster learning trend than APPwt group (M, N); in the testing trail, APPswe (P < 0.01) group had a noticeable shorter latency to escape than APPwt group (O).
Biochemical aggregation assays of Aβ42 peptide
We applied two methods to perform the biochemical aggregation experiments of the Aβ42 peptide. The thioflavin S staining curve showed that the aggregation of the E674Q peptide was slow in the early stage, but once aggregation started, the aggregation speed was accelerated in the late stage, and the aggregation degree was greater than that of the wild type. To further verify the aggregation degree of the two peptides, we observed the microscopic aggregation structure of the two peptides under transmission electron microscopy. The E674Q peptide exhibited higher aggregation than the wild-type peptide, especially the formation of filaments that hinged several fibrils. In general, the results indicate that the E674Q beta-amyloid 42 peptide has different properties from wild-type beta-amyloid with a higher aggregate propensity and slower aggregation speed (Fig. S2).
APPE674Q mutation may develop cognitive deficits and express more C-terminal fragment (CTF) in vivo
After verifying the pathogenicity of APPE674Q in vitro, we tried to verify the pathogenic effect of APPE674Q in vivo. Therefore, we constructed four adeno-associated virus vectors, AAV blank control, AAV-APPwt, AAV-APPE674Q, and AAV-APPswe. Then, we injected the target gene into the CA1 region of the mouse hippocampus through stereotaxic injection so that the related gene could be expressed in the mouse. According to Jaworski's study, related pathological manifestations appeared 6 months after AAV-APP injection in C57BL/6 J mice, and we also performed behavioral and mouse pathological experiments 6 months after the mice were injected. In the novel object recognition test, it is noticeable that the DR2 of APPE674Q animals was less than the blank control (P < 0.05; Fig. 5I, J) suggesting these animals were less inclined to explore and cannot recognize a novel object well. Similar results were found in the Y maze test; APPswe (P < 0.05) and APPE674Q (P < 0.01) had significant differences from the control but no noticeable difference from APPwt (Fig. 5L). However, there were no significant differences among APPswe, APPE674Q, and APPwt in the ratio of DR2/DR1 (P > 0.05; Fig. 5K). No significant difference was found after removing the first day's propensity effect on objects (data not shown). As shown in Figure 5M, the Barnes maze test indicated that APPwt and blank control animals learned the location of the target hole faster than the AAVswe and AAVE674Q groups on Day 5 (P < 0.01). All APP mutation groups showed a potentially faster learning trend than the APPwt group (Fig. 5M, N). In the testing trial, the APPswe (P < 0.01) group had a noticeably shorter latency to escape than the APPwt group, with a 45.99 s advantage. The APPE674Q (P = 0.0555) group also had a possible shorter latency than the APPwt group, with a 32.67 s advantage (Fig. 5O).
Then, we compared the levels of APP and amyloid derivatives in the hippocampi of AAV blank control, AAV-APPwt, AAV-APPE674Q, and AAV-APPswe mice (Fig. 5E, 6 months after injection, i.e., in eight-month-old mice, antibody: Y188, ab32136, Abcam). The total APP was found to be apparently higher in the APPE674Q group (P < 0.001; Fig. 5F) than in the APPwt group, as well as in the AAV-APPswe group compared with the APPwt group (P < 0.0001; Fig. 5F). As observed for the CTF fragment, CTF-β is significantly higher in AAV-APPE674Q than in AAV-APPwt and seems to be as pathogenic as APPswe (P < 0.0001; Fig. 5G). In general, we speculate that the in vivo environment is far more complex than in vitro cells, and the APPE674Q mutation has other crossover pathogenic mechanisms in the mouse model. Under the combined effect of the endogenous APP protein pathologic pathway and AAV human APP pathologic pathway expression, the APP E674Q mutation may promote AD by facilitating the BACE1-mediated processing of APP, resulting in more significant learning and memory behavior impairment. It is also a direction for us to further explore the pathogenesis of APPE674Q by a CRISPR‒Cas9 knock-in mouse model.
Discussion
Here, we report the identification of a novel APP mutation, E674Q, which might be the only pathogenic mutation within the amyloid processing sequence (exons 16–17) to cause LOAD identified thus far. This rare mutation in APP was detected in a late-onset AD pedigree. Symptoms of the proband (Ⅱ-4) appeared in the mid-70s with an insidious onset. Diagnosis of AD was made based on the ATN framework. Next, the amino acid substitution (Glu replaced by Gln in codon 674) was not only indicated to be deleterious by prediction tools and protein simulation analysis but also verified as affecting a functional change in Aβ processing and production at the cellular level. Furthermore, AAV-E674Q-injected mice exhibited impaired learning behavior and increased pathological burden in the brain, implying that the in vivo E674Q mutation had a pathogenicity that bore a comparison with the classical Swedish mutation.
It is worth noting that E674Q is very close to the β-secretase cleavage site and the reported definite pathogenic Swedish mutation (Swedish APP and KM670/671NL).22 No other mutation has been reported for residue 674. Interestingly, two mutations occurring in codon 673 had entirely different phenotypes. APP A673T is the only identified protective APP mutation that reduces the generation of amyloidogenic Aβ peptides by approximately 40% in vitro and protects against cognitive decline in elderly individuals without AD.23,24 The other mutation, APP A673V, was associated with recessive inheritance of AD, which differs from other mutations. To date, a total of 61 APP mutations have been detected worldwide, and ethnic heterogeneity might exist. Few of them are located near the β-secretase cleavage site. In addition to the only protective mutation A673T, other adjacent mutations (A673V, KM670/671NL, and V669L) all lead to EOAD with age at onset ranging from 36 to 61 and progressing to the terminal phase within 4–8 years (Table 1). In contrast, the patient with the E674Q APP mutation and his affected relatives in this study had a more extended duration. The pathogenic mechanism of APP V669L remains unclear, while the Swedish mutation and A673V both increase Aβ production.25 Other mutations within the Aβ sequence might also influence oligomerization kinetics in addition to (or instead of) affecting the amyloidogenic processing of APP.26, 27, 28, 29 Therefore, we performed MD simulations, biochemical aggregation assays, and in vitro cellular experiments to answer the following question: what is the mechanism of molecular pathogenesis in the phenotype of an APP E674Q carrier?
Table 1.
Comparison of probands with APP mutation located nearby β-secretase cleavage site.
| Mutation | E674Q (Shanghai) | A673V | KM670/671NL (Swedish) | V669L (Seoul) |
|---|---|---|---|---|
| Inheritance pattern | Dominant | Recessive | Dominant | Dominant |
| Distance from the cleavage site | +3 | +2 | −2/−1 | −3 |
| Affected first-degree relatives | 1 | 1 | 1–4 | 1 |
| AAO (years) | 70s–80s | 36–46 | 45–61 | 56, 50s |
| Duration of progression | Scores of MMSE dropped to 17 in 3 years | 7 years on average | scores of MMSE dropped from 25 to 13 in one year; 4 years to become mute and bedridden after the 1st symptom | |
| Non-cognitive symptoms | Not informed | Myoclonic jerks | NA | Bradykinesia, limb rigidity, short-stepped gait, masked face; myoclonic jerks, seizure |
| Reference | 33 | 22 | 34 |
Note: Distance from the cleavage site is calculated from Asp672 (Asp +1). NA, not available.
To explore APP-induced conformational switching of BACE1, we performed MD simulations. By integrating results from a series of computational techniques, a possible cleavage process in the E674Q mutant system can be proposed (Fig. 4F). When the E674Q APP fragment gets close to the catalytic cavity of BACE1, electrostatic interaction is the driving force for their binding and subsequent catalysis. E674Q mutation of APP resulted in the change of the interaction mode between APP and BACE1. The flap of BACE1 in the E674Q system of APP and BACE1 flipped up, and BACE1 was in an “open state”, while it flipped down in the WT APP-BACE1 system, wherein BACE1 was in a “closed state”. As a result, the catalytic dyad in the E674Q system got close to the cleavage site of APP, while it was far in the WT system. Therefore, the E674Q mutant APP is likely more prone to be cleaved by BACE1, which may subsequently promote the aggregation of Aβ.
As is well-known, APP undergoes amyloidogenic and non-amyloidogenic processing. To produce Aβ, APP is first cleaved by BACE1, and then by γ-secretase. As above-mentioned, several APP mutations adjacent to the BACE1 cleavage site, such as KM670/671NL (Swedish mutation) and D678N, have been reported to enhance the BACE1-mediated cleavage of APP. We found that the subject with the E674Q mutation showed Aβ deposition in the brain by 18F-florbetapir PET imaging. Thus, we further tested the effect of the APP E674Q mutation in vitro. Results revealed that the E674Q substitution substantially increased the processing of APP by BACE1, as shown by the production of C99. The effect of E674Q on the production of C99 and Aβ was even more severe than in cells transfected with the well-recognized APPswe mutation. Considering that the condition of the patient with APP E674Q was not that aggressive compared to individuals with APPswe mutation, this might indicate that symptoms of AD are not completely determined by Aβ. To date, evidence has shown that tau neurofibrillary pathology and other pathology have a stronger connection with cognition.30 Interestingly, the A673T mutation, which occurs on the residue adjacent to E674 protects against AD.24 It is conceivable that the APP mutations near the BACE1-mediated cleavage site alter the conformation of APP, and affect the interaction between BACE1 and APP. This ultimately could promote the production of Aβ as seen with the Swedish mutation, D678N, and the E674Q mutation, or it can inhibit the production of Aβ as with the A673T mutation.
Meanwhile, we also noticed the question that the patient with E674Q had relatively late-onset symptoms, in contrast to patients with the Swedish mutation. We made a synthetic beta-amyloid peptide carrying the E674Q mutation and performed biochemical aggregation experiments. Interestingly, the E674Q peptide exhibited higher aggregation compared with the wildtype peptide, especially the formation of filaments that hinged several fibrils. In general, the E674Q Aβ42 peptide has different properties from wildtype beta-amyloid with a higher aggregate propensity and slower aggregation speed, which could partly explain the E674Q (“Shanghai APP”) in a Chinese patient with typical LOAD, rather than EOAD (Fig. S2).
Furthermore, we also built an APPE674Q mouse model by AAV injection. After 6 months, behavioral tests showed that the APPE674Q mutation may lead to cognitive deficits in vivo. Consistently, the hippocampal Western blot results corresponded with the behavior test results. Due to the limitations of AAV infection efficiency and presentation duration, this model does not fully demonstrate its pathogenic effect. In the future, it is still necessary to build more disease models that are more like the human environment through methods such as gene editing.
To our knowledge, E674Q might be the only pathogenic mutation within the amyloid processing sequence (exons 16–17) to cause LOAD. LOAD is believed to be controlled by a group of common risk alleles across multiple genes. Genetic modifiers have a significant impact on disease susceptibility and age of onset.31 Previously, a few other mutations in APP were detected in patients with LOAD, located in exons 5, 13, and 14, outside the Aβ sequence, and confirmed to be likely benign polymorphisms.32 Given the lack of samples from other relatives in the present study, the penetrance rate and average age at onset for AD in APP E674Q carriers remain unclear. Additionally, the study subject's son (III-1) as an E674Q carrier needs to receive follow-up with cognitive testing.
In conclusion, we present a novel missense APP mutation, E674Q (Shanghai APP), in a Chinese patient with typical LOAD and probable positive family history. The amino acid replacement located near the β-secretase cleavage site was predicted to be damaging or disease-causing in silico. Energy-minimized protein structure analysis showed that the mutation would cause BACE1 to be in a more open active conformation than that which bound with WT APP substrate, and the efficiency of enzymatic cleavage increased in vitro with the APP E674Q mutation. Increased accumulation subsequently promotes the aggregation of Aβ. In this study, we find that the E674Q mutation is potentially disease-causing by facilitating the BACE1-mediated processing of APP and the production of Aβ, providing insight into the molecular structure–function relationship of the APP-BACE1 enzyme–substrate complex.
Conflict of interests
The authors have declared no conflict of interests.
Funding
This study was supported by grants from the Ministry of Science and Technology of the People's Republic of China (No. 2021ZD020180) and the Natural Science Foundation of China (No. 81971068, 81922021, 81773635, 82073765).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2023.02.051.
Contributor Information
Zhentao Zhang, Email: zhentaozhang@whu.edu.cn.
Wei Fu, Email: wfu@fudan.edu.cn.
Gang Wang, Email: wgneuron@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Fig. S1.
The time-evolved potential energy and RMSD of MD systems. (A) The time-evolved potential energy of the apo system. (B) The time-evolved potential energy for BACE1 in the four systems. (C) The time-evolved RMSD values for BACE1 in the four systems.
Fig. S2.
Biochemical aggregation of Amyloid-beta 42 systems. (A) Transmission electron microscopy (TEM) micrograph of wild-type Amyloid-beta 42 (Aβ42) fibrils. (B) TEM micrograph of the E674Q Aβ42 fibrils. (C) Thioflavin-S staining characterizes Aβ42 aggregation.
References
- 1.2020 Alzheimer's disease facts and figures. Alzheimers Dement. 2020;16:391–460. [Google Scholar]
- 2.van der Flier W.M., AL Pijnenburg Y., Fox N.C., et al. Early-onset versus late-onset Alzheimer's disease: the case of the missing APOE ϵ4 allele. Lancet Neurol. 2011;10(3):280–288. doi: 10.1016/S1474-4422(10)70306-9. [DOI] [PubMed] [Google Scholar]
- 3.Tang M., Ryman D.C., McDade E., et al. Neurological manifestations of autosomal dominant familial Alzheimer's disease: a comparison of the published literature with the Dominantly Inherited Alzheimer Network observational study (DIAN-OBS) Lancet Neurol. 2016;15(13):1317–1325. doi: 10.1016/S1474-4422(16)30229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen J.C., Beck J.A., Campbell T.A., et al. Early onset familial Alzheimer's disease: mutation frequency in 31 families. Neurology. 2003;60(2):235–239. doi: 10.1212/01.wnl.0000042088.22694.e3. [DOI] [PubMed] [Google Scholar]
- 5.Blauwendraat C., Wilke C., Jansen I.E., et al. Pilot whole-exome sequencing of a German early-onset Alzheimer's disease cohort reveals a substantial frequency of PSEN2 variants. Neurobiol Aging. 2016;37:208.e11–208.e17. doi: 10.1016/j.neurobiolaging.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Giau V.V., Bagyinszky E., Youn Y.C., et al. APP, PSEN1, and PSEN2 mutations in Asian patients with early-onset Alzheimer disease. Int J Mol Sci. 2019;20(19):4757. doi: 10.3390/ijms20194757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y., Ren R.J., Zhong Z.L., et al. Mutation profile of APP, PSEN1, and PSEN2 in Chinese familial Alzheimer's disease. Neurobiol Aging. 2019;77:154–157. doi: 10.1016/j.neurobiolaging.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Jia L., Fu Y., Shen L., et al. PSEN1, PSEN2, and APP mutations in 404 Chinese pedigrees with familial Alzheimer's disease. Alzheimers Dement. 2020;16(1):178–191. doi: 10.1002/alz.12005. [DOI] [PubMed] [Google Scholar]
- 9.Lanoiselée H.M., Nicolas G., Wallon D., et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: a genetic screening study of familial and sporadic cases. PLoS Med. 2017;14(3) doi: 10.1371/journal.pmed.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cacace R., Sleegers K., Van Broeckhoven C. Molecular genetics of early-onset Alzheimer's disease revisited. Alzheimers Dement. 2016;12(6):733–748. doi: 10.1016/j.jalz.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Karch C., Cruchaga C., Goate A. Alzheimer's disease genetics: from the bench to the clinic. Neuron. 2014;83(1):11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKhann G.M., Knopman D.S., Chertkow H., et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie X.Y., Zhao Q.H., Huang Q., et al. Genetic profiles of familial late-onset Alzheimer's disease in China: the Shanghai FLOAD study. Genes Dis. 2022;9(6):1639–1649. doi: 10.1016/j.gendis.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urban A.S., Pavlov K.V., Kamynina A.V., et al. Structural studies providing insights into production and conformational behavior of amyloid-β peptide associated with Alzheimer's disease development. Molecules. 2021;26(10):2897. doi: 10.3390/molecules26102897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raveh B., London N., Schueler-Furman O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins. 2010;78(9):2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 16.London N., Raveh B., Cohen E., et al. Rosetta FlexPepDock web server: high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39:W249–W253. doi: 10.1093/nar/gkr431. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Meisl G., Frohm B., et al. On the role of sidechain size and charge in the aggregation of Aβ42 with familial mutations. Proc Natl Acad Sci U S A. 2018;115(26):E5849–E5858. doi: 10.1073/pnas.1803539115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheltens P., Leys D., Barkhof F., et al. Atrophy of medial temporal lobes on MRI in "probable" Alzheimer's disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992;55(10):967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerreiro R.J., Baquero M., Blesa R., et al. Genetic screening of Alzheimer's disease genes in Iberian and African samples yields novel mutations in presenilins and APP. Neurobiol Aging. 2010;31(5):725–731. doi: 10.1016/j.neurobiolaging.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R., Lai L., Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 21.Cole S.L., Vassar R. The Alzheimer's disease beta-secretase enzyme, BACE1. Mol Neurodegener. 2007;2:22. doi: 10.1186/1750-1326-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullan M., Crawford F., Axelman K., et al. A pathogenic mutation for probable Alzheimer's disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1(5):345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 23.Peacock M.L., Warren J.T., Jr., Roses A.D., et al. Novel polymorphism in the A4 region of the amyloid precursor protein gene in a patient without Alzheimer's disease. Neurology. 1993;43(6):1254–1256. doi: 10.1212/wnl.43.6.1254. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson T., Atwal J.K., Steinberg S., et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 25.Yan R., Vassar R. Targeting the β secretase BACE1 for Alzheimer's disease therapy. Lancet Neurol. 2014;13(3):319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakutani Y., Watanabe K., Adachi Y., et al. Novel amyloid precursor protein gene missense mutation (D678N) in probable familial Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2004;75(7):1039–1042. doi: 10.1136/jnnp.2003.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W.T., Hong C.J., Lin Y.T., et al. Amyloid-beta (Aβ) D7H mutation increases oligomeric Aβ42 and alters properties of Aβ-zinc/copper assemblies. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y.C., Wang J.Y., Wang K.C., et al. Differential regulation of amyloid precursor protein sorting with pathological mutations results in a distinct effect on amyloid-β production. J Neurochem. 2014;131(4):407–412. doi: 10.1111/jnc.12829. [DOI] [PubMed] [Google Scholar]
- 29.Li N.M., Liu K.F., Qiu Y.J., et al. Mutations of beta-amyloid precursor protein alter the consequence of Alzheimer's disease pathogenesis. Neural Regen Res. 2019;14(4):658–665. doi: 10.4103/1673-5374.247469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagust W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat Rev Neurosci. 2018;19(11):687–700. doi: 10.1038/s41583-018-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milind N., Preuss C., Haber A., et al. Transcriptomic stratification of late-onset Alzheimer's cases reveals novel genetic modifiers of disease pathology. PLoS Genet. 2020;16(6) doi: 10.1371/journal.pgen.1008775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassi C., Guerreiro R., Gibbs R., et al. Investigating the role of rare coding variability in Mendelian dementia genes (APP, PSEN1, PSEN2, GRN, MAPT, and PRNP) in late-onset Alzheimer’s disease. Neurobiol Aging. 2014;35(12):2881.e1–2881.e6. doi: 10.1016/j.neurobiolaging.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Fede G., Catania M., Morbin M., et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science. 2009;323(5920):1473–1477. doi: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagyinszky E., Kang M.J., Van Giau V., et al. Novel amyloid precursor protein mutation, Val669Leu (“Seoul APP”), in a Korean patient with early-onset Alzheimer’s disease. Neurobiol Aging. 2019;84:236.e1–236.e7. doi: 10.1016/j.neurobiolaging.2019.08.026. [DOI] [PubMed] [Google Scholar]