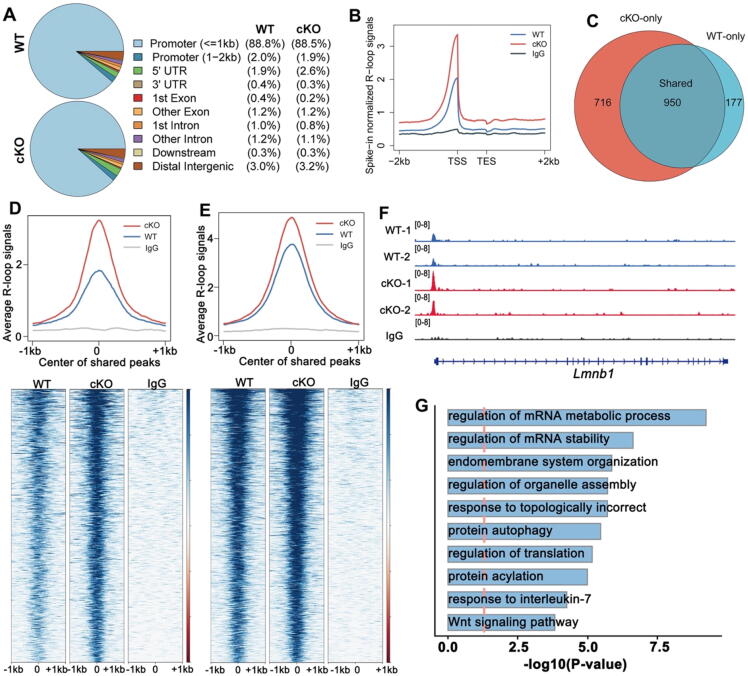
Fig. 5.
R-loops accumulate in Rnaseh1fl/−;Stra8-Cre spermatocytes. A: Pie charts showing the genomic distribution of R-loops in leptotene/zygotene spermatocytes of WT and Rnaseh1fl/−;Stra8-Cre mice.B: Genomic metaplots showing relative signal intensity comparison of R-loops from the transcription start sites (TSSs) to transcription end sites (TESs) in WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes. C: Venn diagram showing the overlap of R-loop peaks in WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes (Fisher’s exact test; ***P < 0.001). D: Average R-loop read density and heatmap of WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes in the 1 kb window around the center of cKO-only peaks (D) and shared R-loop peaks. Loci are ordered by the descending R-loop intensities in cKO samples. E: Average R-loop read density and heatmap of WT and Rnaseh1fl/−;Stra8-Cre leptotene/zygotene spermatocytes in the 1 kb window around the center of shared R-loop peaks. Loci are ordered by the descending R-loop intensities in cKO samples. F: Snapshots of accumulated R-loops on example genes. G: Gene ontology analysis of R-loop accumulated genes in Rnaseh1-null spermatocytes.