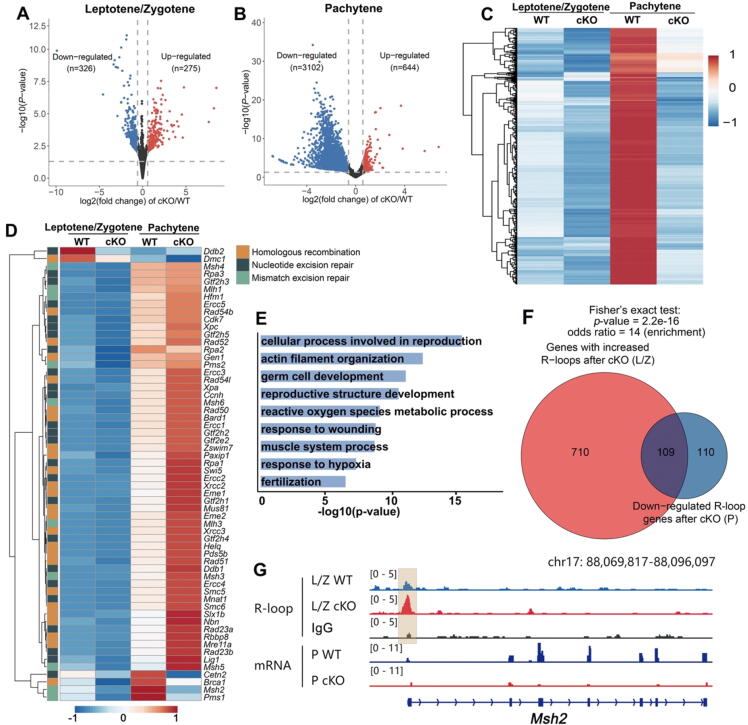
Fig. 6.
RNA-seq analyses of leptotene/zygotene and pachytene spermatocytes derived from WT and Rnaseh1fl/−;Stra8-Cre mice. A-B: Volcano plots showing differentially expressed genes of leptotene/zygotene and pachytene spermatocytes. Genes upregulated (FC > 2; FDR < 0.05) and downregulated (FC < 1/2; FDR < 0.05) in spermatocytes are colored red and blue, respectively. C: Dynamic profiling of the 3,102 downregulated genes in Rnaseh1fl/−;Stra8-Cre pachytene spermatocytes. D: Dynamic profiling of DNA damage repair and homologous recombination-related genes across different stages in WT and Rnaseh1fl/−;Stra8-Cre spermatocytes. E: Gene ontology analysis of differentially expressed genes in Rnaseh1-null pachytene spermatocytes. F: Venn diagram showing the overlap of genes with accumulated R-loops in Rnaseh1-null leptotene/zygotene spermatocytes and down-regulated R-loop genes in pachytene spermatocytes. L/Z, leptotene/zygotene; P, pachytene. G: Example snapshots of the increased R-loops and downregulated transcription in Rnaseh1-null spermatocytes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)