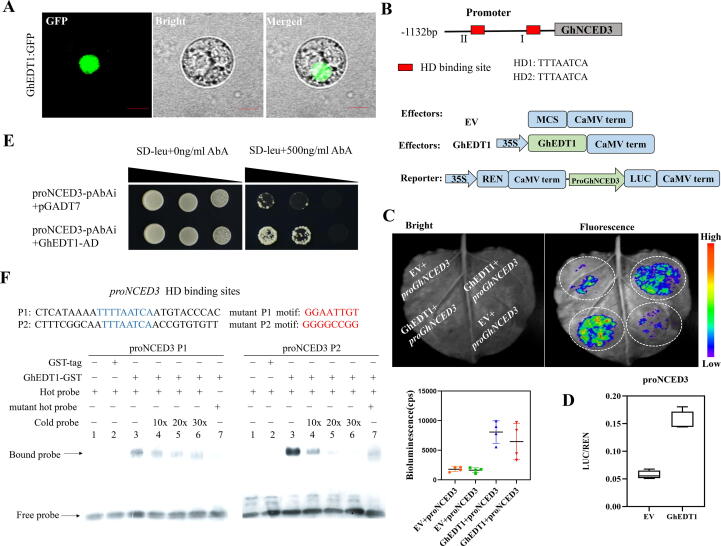
Fig. 6.
GhEDT1 binds to the promoter of GhNCED3 to activate its expression and promote ABA synthesis. (A) Subcellular localization of GhEDT1 in cotton protoplasts. Bars = 10 μm. (B) Distribution diagram of HD binding sites contained in GhNCED3 promoter region. Schematic diagram of effector and reporter constructs. The empty vector (EV) was used as a control. LUC signals are shown in the image. (C) The intensity of fluorescence in dual-luciferase reporter assays. The bioluminescence reflects the trans-activation ability of GhEDT1. (D) The transient dual-luciferase reporter assays in cotton (YZ1) protoplasts. The ratio of firefly luciferase (LUC) to renilla luciferase (REN) reflects the trans-activation ability of GhEDT1. (E) Yeast-one-hybrid assays revealed that GhEDT1 could bind the GhNCED3 promoter region. The target fragment containing two HD binding sites was cloned into the pAbAi vector, and the full-length coding sequence (CDS) of GhEDT1 was fused to the pGADT7 vector. Interaction was determined on selective medium lacking Leu in the presence of 500 ng ml−1 aureobasidin A (SD/−Leu + AbA500). Different colony dots of the yeast represent different dilution times. (F) EMSA assay of the DNA binding activity of GhEDT1 for the GhNCED3 promoter. P1 and P2 probes were labelled with biotin as hot probes and incubated with recombinant GhEDT1-GST protein. Unlabeled probes as cold probes were added to compete with biotin-labelled probe. Mutation probes as control. GST protein was used as negative control. Values represent means ± SE (n = 3).