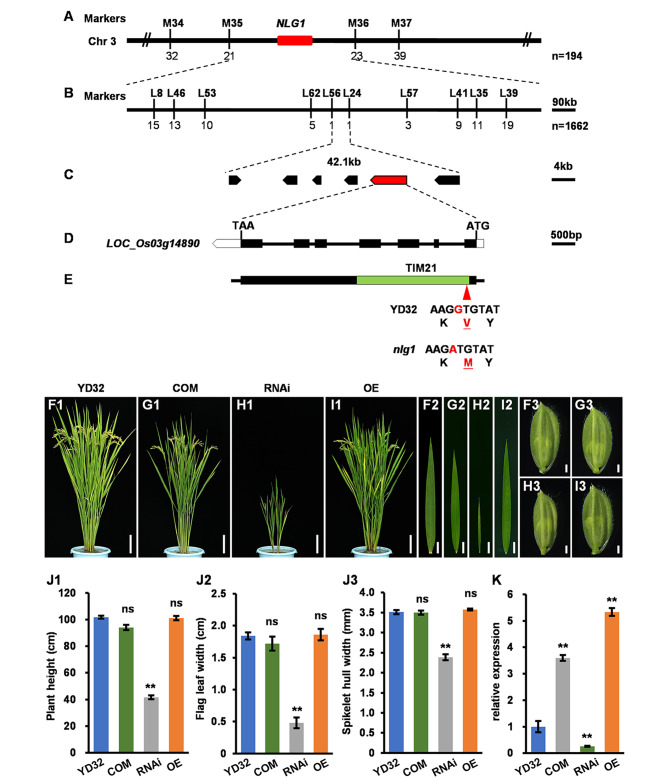
Fig. 4.
Map-based cloning and functional confirmation of NLG1. A-C Mapping of NLG1 in a 42.1-kb region on chromosome 3. The numbers below the markers represents the recombinants. D-E A single base-pair substitution from G to A occurred in seventh exon of LOC_Os03g14890, leading to a residue alteration from Val to Met in TIM21 domain. F-I Morphological comparison of plant architectures, flag leaves and spikelet hulls of YD32 (F1, F2 and F3), COM (NLG1-complementation) (G1, G2 and G3), RNAi (NLG1-RNA interference) (H1, H2 and H3), OE (NLG1-overexpression) (I1, I2 and I3) transgenic lines. J1-J3 Plant height (J1), flag leaf width (J2) and spikelet hull width (J3) of YD32, COM, RNAi and OE transgenic lines. Data represent means ± SD (n = 5). K Expression analysis of NLG1 in the flag leaves of the YD32, COM, RNAi and OE transgenic lines using RT-qPCR. Data represent means ± SD (n = 3). **Significant difference at p < 0.01 compared with YD32 by Student’s t-test, and ns means no significance. Scale bars: 10 cm in F1-I1, 2 cm in F2-I2, 1 mm in F3-I3