Figure 3. Genomic analysis of experimentally evolved isolates reveals molecular basis for mucin-mediated aggregation defects.
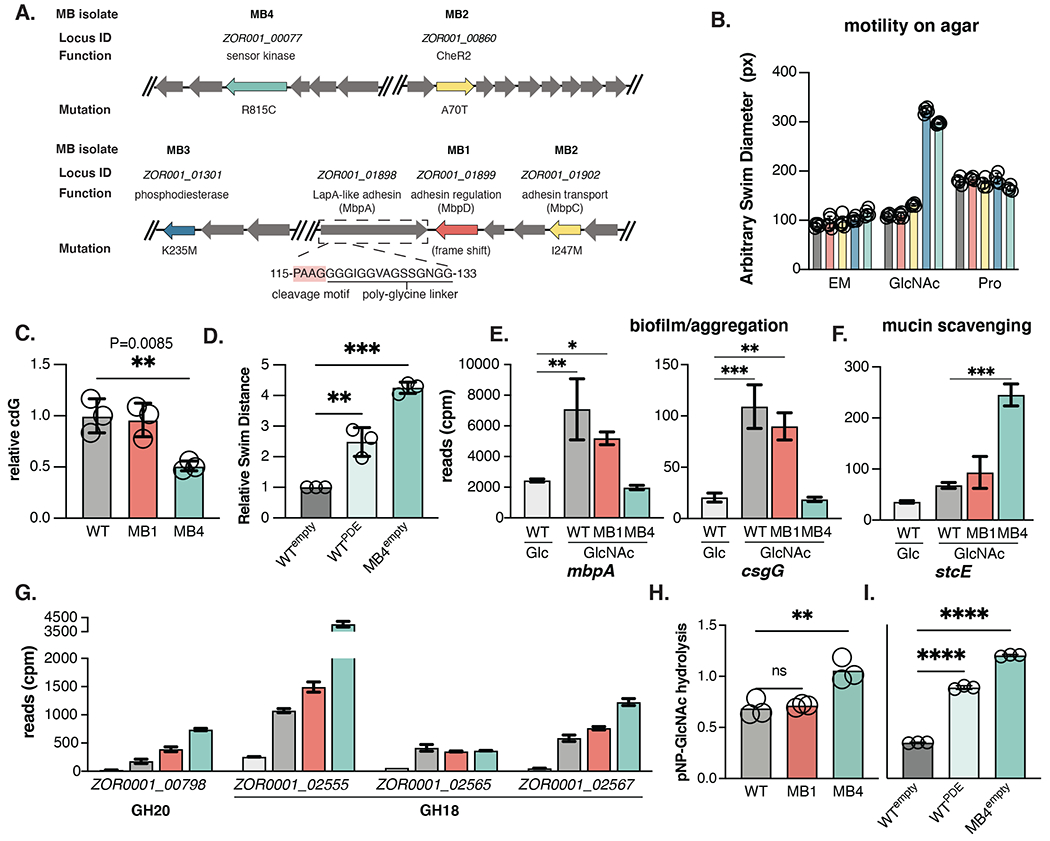
(A) Chromosomal organization of MB1-MB4 mutations (red: MB1; yellow: MB2; blue: MB3; teal: MB4). The locus ID, putative function, nucleotide substation, and corresponding amino acid change listed when applicable. The ORF encoding mbpA is boxed and the amino acids corresponding to the MbpG cleavage motif in the N-terminal retention module are indicated.
(B) Swim diameter of WT and MB isolates in MM alone (N=5 replicates) or supplemented with 0.4% GlcNAc (N= 5) or 1 mM proline (N=4) after 20 hr incubation at 30°C.
(C) Quantification of c-di-GMP in WT, MB1, and MB4 exposed to GlcNAc.
(D) Relative swim distance of Aer01 engineered to express the PDE ZOR0001_03617 upon aTc induction compared to Aer01 and MB4 carrying an empty induction construct.
(E-G) RNAseq analysis of indicated genes involved in biofilm formation/aggregation and mucin scavenging following exposure to 0.4% Glucose (WT, N=3) or 0.4% GlcNAc (WT, N=3; MB1, N=3; MB4, N=2). Ordinary one-way ANOVA with multiple comparisons. See also Table S1.
(H) Hydrolytic activity of WT, MB1, and MB4 on pNP-GlcNAc (N=3).
(I) Suppressing c-di-GMP accumulation increases Aer01 pNP-GlcNAc hydrolysis (N=3). Mean and standard deviations are indicated in all graphs.
