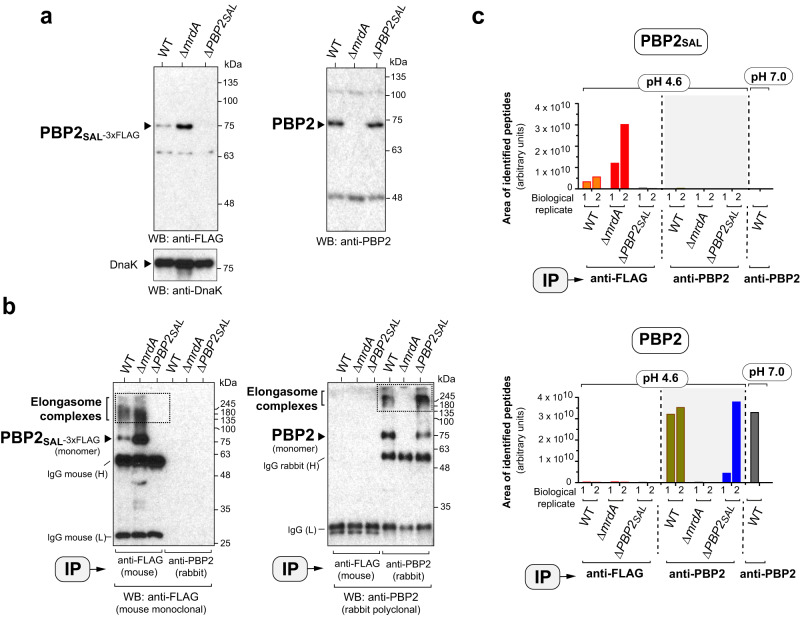
Fig. 5. PBP2SAL and PBP2 assemble in independent elongasomes.
a Control Western assays showing the specificity of the anti-FLAG and anti-PBP2 antibodies used to pull down the PBP2SAL- and PBP2 elongasomes. Shown are total protein fractions of the indicated strains grown to exponential phase (OD600 ~0.5) in LB pH 4.6. The unrelated protein DnaK was detected in this input material as loading control. Positions and sizes of the molecular weight markers (in kDa) are indicated. b Pulldown of the PBP2SAL- and PBP2-elongasomes from membrane fractions of the indicated strains. Wild-type (WT) and ΔmrdA cells bear an 3xFLAG-epitope tag at the 3’-end of the gene encoding PBP2SAL in its native chromosomal position. Bacteria were grown in LB pH 4.6, washed, and incubated for 30 min at room temperature (RT) in PBS pH 8.0 buffer containing the cross-linker reagent DSS (disuccinimidyl suberate). The cross-linking reaction was quenched with 20 mM Tris for 15 min at RT. Note the existence of independent elongasomes. In one case, the presence of PBP2SAL-3xFLAG in high-molecular complexes cross-linked with DSS in WT and ΔmrdA cells. In the other case, high-molecular weight complexes pulled down with anti-PBP2 antibody are only visible in WT and ΔPBP2SAL cells (n = 2 biological). Bands corresponding to the heavy (H) and light (L) of the primary monoclonal mouse antibody anti-FLAG are indicated. IP, antibody used in the immunoprecipitation. Positions of the elongasome complexes and sizes of the molecular weight markers (in kDa) are also indicated. c Sum of the integrated areas of the peaks corresponding to the set of peptides identified for PBP2SAL or PBP2 (see “Supplementary Data” file) in the complexes immunoprecipitated (IP) from membrane fractions of bacteria grown at the indicated pH with anti-FLAG (PBP2SAL) or anti-PBP2 antibodies (n = 2 biological). In the neutral pH condition, the pull down was performed exclusively with anti-PBP2 antibody given the no production of PBP2SAL by WT bacteria in this condition (see Fig. 1c). The proteomic data were processed using the Skyline v22.2 software (https://skyline.ms/project/home/software/Skyline/begin.view).