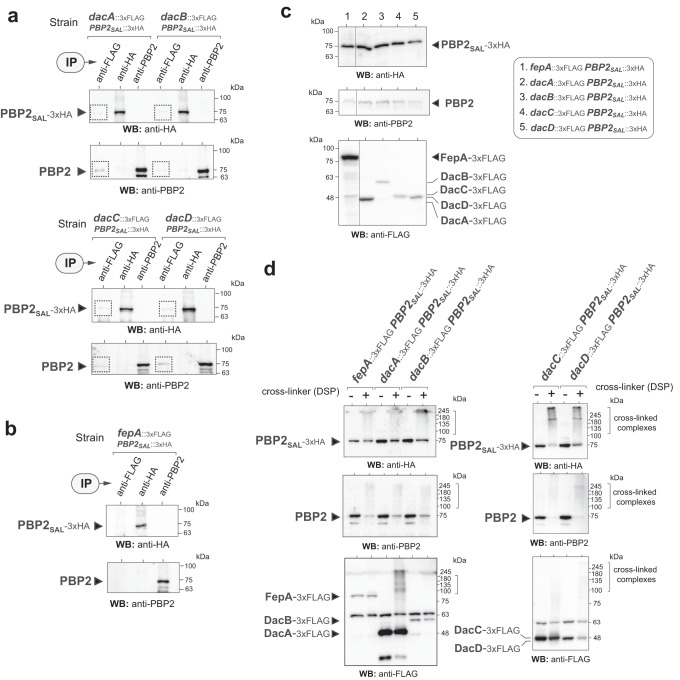
Fig. 6. The D,D-CPases DacA, DacB, DacC and DacD are present in a fraction of PBP2- and PBP2SAL-elongasomes.
a Immuno-precipitates of elongasome complexes from wild-type cells grown in LB pH 4.6. The isogenic strains used for these assays, all in wild-type genetic background and bearing a 3xHA epitope tag in the 3’ end of the PBP2SAL-encoding gene, bear an 3xFLAG epitope in the respective genes encoding D,D-CPases (dacA, dacB, dacC, dacD) (n = 2 biological). Complexes were cross-linked in vivo with the reagent DSP (dithiobis[succinimidylpropionate]), immunoprecipitated (IP) with the indicated antibodies and the gels run in denaturing conditions, containing β-mercaptoethanol. The presence of PBP2SAL or PBP2 in complexes pulled down with the anti-FLAG antibodies recognising D,D-CPases is highlighted with boxes (n = 2 biological). IP, antibody used in the immunoprecipitation. Positions and sizes of the molecular weight standards (in kDa) are indicated. b Control immunoprecipitation assays in a strain bearing a 3xFLAG epitope in the unrelated inner membrane FepA, involved in iron transport (n = 2 biological). Positions and sizes of molecular weight standards (in kDa) are indicated. c Relative levels in the input samples of the proteins under study (PBP2SAL, PBP2, DacA, DacB, DacC, DacD, FepA) in the membrane extracts prepared prior to the in vivo cross-linking (n = 2 biological). Two different areas of the blots were spliced together as indicated by the line shown between lines 1 and 2 of the blots. Positions and sizes of molecular weight standards (in kDa) are indicated. d Presence of the proteins under analysis (PBP2SAL, PBP2, DacA, DacB, DacC, DacD, FepA) in higher molecular weight complexes following cross-linking with DSP. Samples are compared in bacteria exposed or not to cross-linker and the gels run in the absence of β-mercaptoethanol (n = 2 biological). Positions and sizes of molecular weight standards (in kDa) are indicated.