Abstract
To effectively track and eliminate COVID-19, it is critical to develop tools for rapid and accessible diagnosis of actively infected individuals. Here, we introduce a single-walled carbon nanotube (SWCNT)-based optical sensing approach toward this end. We construct a nanosensor based on SWCNTs noncovalently functionalized with ACE2, a host protein with high binding affinity for the SARS-CoV-2 spike protein. The presence of the SARS-CoV-2 spike protein elicits a robust, 2-fold nanosensor fluorescence increase within 90 min of spike protein exposure. We characterize the nanosensor stability and sensing mechanism and passivate the nanosensor to preserve sensing response in saliva and viral transport medium. We further demonstrate that these ACE2-SWCNT nanosensors retain sensing capacity in a surface-immobilized format, exhibiting a 73% fluorescence turn-on response within 5 s of exposure to 35 mg/L SARS-CoV-2 virus-like particles. Our data demonstrate that ACE2-SWCNT nanosensors can be developed into an optical tool for rapid SARS-CoV-2 detection.
Keywords: SARS-CoV-2, nanosensor, protein detection, carbon nanotube, fluorescence
Graphical Abstract
The World Health Organization deemed COVID-19 a global pandemic on March 11, 2020. As of February 14, 2021, SARS-CoV-2 has infected over 108 million people and caused over 2.3 million deaths worldwide.1 It is estimated that over 70% of infected individuals under the age of 60 are asymptomatic yet can still transmit the virus to others.2 Early estimates placed the basic reproductive number (R0) at 2.2, which represents the average number of people an infected person will spread the disease to.3 Taken together, these findings underscore the need for advancements in testing and containment efforts to end the pandemic.
Current SARS-CoV-2 testing strategies can be grouped into two categories: molecular tests and serological tests. Molecular tests remain the status quo for diagnosing active CoV-2 infections by detecting CoV-2 RNA in patient samples, including sputum and nasal fluid. Molecular tests primarily use real-time reverse transcription polymerase chain reaction (RT-PCR) to amplify and detect CoV-2 RNA, a process that is expensive ($5–10 per test), time-consuming (2–3 h), and requires laboratory processing.4–7 Yet, RT-PCR tests possess high sensitivity in identifying viral nucleic material, with the limit of detection (LOD) reported between 1 and 10 viral RNA copies necessary to produce a positive result.4 Serological tests detect the presence of IgG and IgM antibodies in patient blood serum and provide important surveillance data of past viral infections, though do not identify active cases. As such, detection of viral RNA by molecular tests is to-date the preferred testing mode to diagnose active CoV-2 cases. However, the complexity of the process necessitates the use of expensive equipment and trained personnel, limiting the testing capability of rural and lower income regions.8 Altogether, these factors amount to a large enough backlog in RT-PCR testing capabilities such that the United States is at 52% of its daily testing target to mitigate the spread of the virus as of November 1, 2020.9 Several non-PCR-based methods of viral RNA detection have been developed recently, implementing techniques such as loop-mediated isothermal amplification (LAMP),10 localized surface plasmon resonance (LSPR),11 and CRISPR machinery12 to avoid the expensive equipment required for the heating and cooling cycles of RT-PCR. However, these techniques are not as sensitive as RT-PCR and still require between 30 min to 1 h of processing time per sample.13 Antigen testing has emerged with great potential for rapid diagnostics, possessing a key strength that active virus is detected. This contrasts with RT-PCR tests, which are merely detecting the presence of viral RNA and can consequently lead to cases of RT-PCR positivity in the absence of any viable virus.14 Although antigen tests are faster and cheaper, they possess lower sensitivity.14
There has been a strong drive to find other viral testing targets and methodologies for simpler and faster diagnostics due to extended processing time and equipment restrictions associated with viral RNA detection. SARS-CoV-2 is a coronavirus, a family of viruses termed as such due to the halo or “corona” of proteins surrounding the virus. These outwardly protruding spike (S) proteins bind to the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of human respiratory endothelial cells, facilitating viral entry.15,16 The viral S protein is also the primary antigen that human monoclonal antibodies bind to prevent host cell entry and mark the virus for clearance.17 With approximately 100 S protein trimers per SARS-CoV-2 virion, the S protein is a prime target for live virus detection.18 For example, Seo et al. developed a field-effect transistor-based sensor by functionalizing graphene sheets with SARS-CoV-2 S protein antibodies to detect SARS-CoV-2 at a LOD of 242 copies/mL in crude, nasopharyngeal swab clinical samples.19 Several promising nanotechnology-based sensors for SARS-CoV-2 detection have also emerged for both nucleic acid- and antigen-based detection and diagnosis of COVID-19, including platforms based on gold nanoparticles and quantum dots.11,20–23 Such technologies will be crucial in working toward sensitive tests that do not rely on specialized equipment for signal readout and controlled laboratory environments for sample processing.13
Single-walled carbon nanotubes (SWCNTs) have shown much utility for biological analyte sensing.24–26 SWCNTs are intrinsically near-infrared (nIR) fluorescent and can be functionalized with various sensing moieties to develop stable biological sensors with rapid fluorescence-change readouts. Unlike conventional fluorophores, SWCNTs do not photo-bleach, giving rise to their potential long-term use.25 Importantly, the SWCNT near-infrared emission is minimally absorbed and scattered by biomolecules,25 providing a readout that can penetrate optically occluded patient samples, thus eliminating the need for sample purification that limits the throughput of other viral testing modes. Furthermore, SWCNTs offer facile incorporation into portable form factors such as immobilization in paper or hydrogels27,28 with detection of the nIR SWCNT signal by a Raspberry Pi and charge-coupled device (CCD) camera system, of similar form factor to a smartphone.29
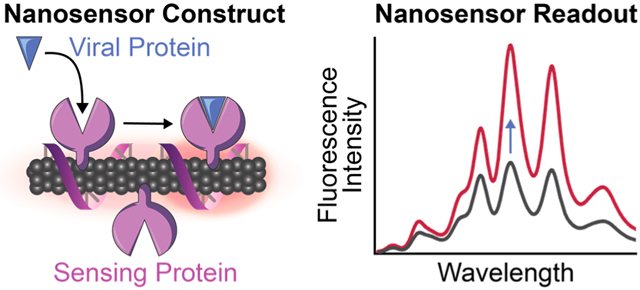
Herein we demonstrate the development and characterization of a nanosensor to detect SARS-CoV-2 by exploiting the innate ability of host proteins to bind virion components, coupled to a SWCNT substrate that provides a fluorescence readout of the protein recognition events. This concept of a hybrid nano-bio sensor harnesses the natural recognition abilities of proteins acting as sensing moieties, together with SWCNTs as signal transducers, to enable targeted biological sensing. Considering the difficulties of preserving protein activity once tethered to nanomaterials, few previous protein-SWCNT conjugate sensors have been developed, and most have used enzymes to detect their small molecule substrates30–32 or protein A to detect antibodies.33,34 Here, we introduce a protein-SWCNT construct that uses human host cell membrane protein ACE2 to bind to the CoV-2 spike protein receptor-binding domain (S RBD) protruding from the virion surface, enabling protein detection in a rapid, label-free manner. We constructed nanosensors by immobilizing ACE2 proteins on the surface of SWCNTs, where this noncovalent modification strategy is advantageous in retaining the intact SWCNT surface lattice that is necessary for fluorescence.30,35 Upon S protein binding to ACE2-functionalized SWCNTs, the change in exciton dynamics of the SWCNTs leads to a modulation in the nIR SWCNT fluorescence. We demonstrate that ACE2-functionalized SWCNT nanosensors can achieve a LOD of 12.6 nM S RBD, can be passivated for detection of S protein in saliva and viral transport medium, and can be imaged for rapid detection of S protein and virus-like CoV-2 virions within seconds.
NANOSENSOR PLATFORM GENERATION AND CHARACTERIZATION
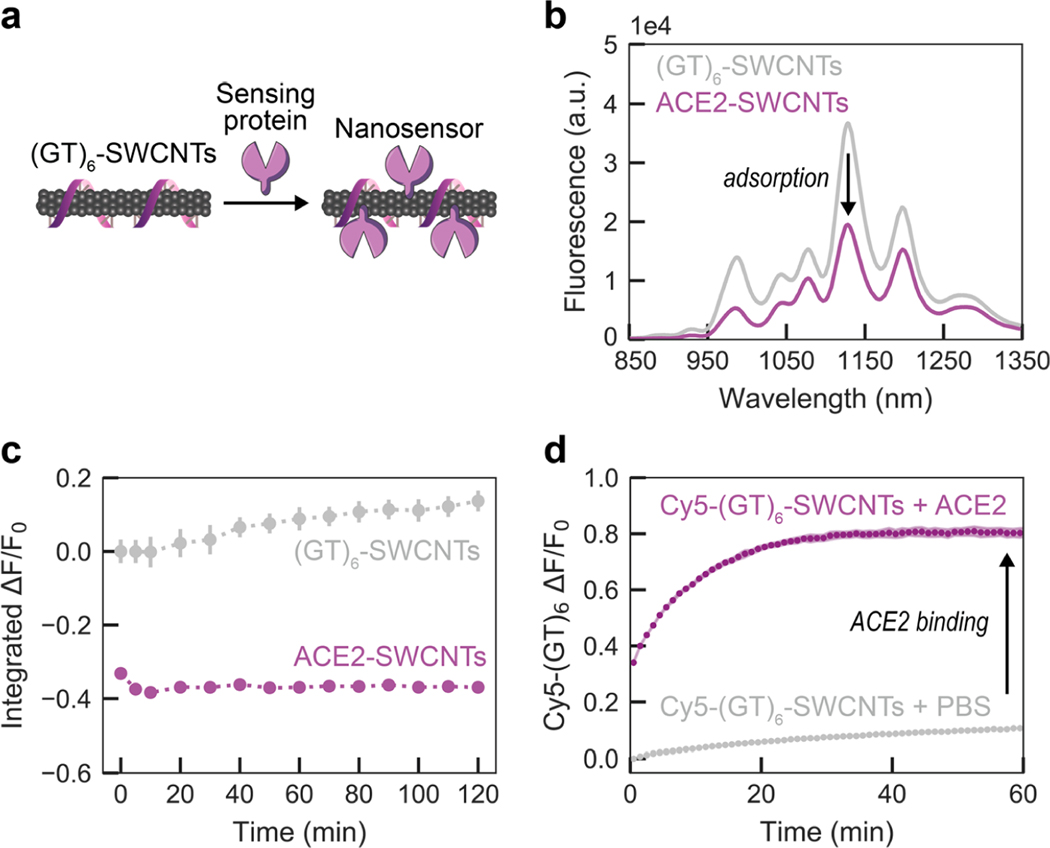
To generate nanosensors, we first solubilized SWCNTs by probe-tip sonication with single-stranded DNA (ssDNA), (GT)6 (see Methods in the Supporting Information). Direct probe-tip sonication of ACE2 with pristine SWCNTs did not lead to a stable suspension and further raises the likelihood for disruption of the native ACE2 protein conformation, and hence loss of sensing ability for S RBD. The ssDNA sequence of guanine-thymine (GT) was chosen on the basis of high SWCNT suspension yield. The short ssDNA sequence length (12 nucleotides) was informed by previous work demonstrating that shorter ssDNA desorbs faster, and to a greater extent, from SWCNTs in the presence of proteins.36,37 (GT)6-SWCNTs (2.5 mg/L final concentration) were incubated with ACE2 sensing protein (6.25 mg/L final concentration) in phosphate-buffered saline (PBS) solution to noncovalently passivate the SWCNT surface with protein, schematically represented inFigure 1a.37 This ratio of ACE2 sensing protein to SWCNT substrate was calculated to be approximately above the close-packing threshold to minimize protein surface-denaturation and colloidal aggregation; then this calculated value was experimentally optimized (see details in the Supporting Information and Figures S1 and S2). ACE2 adsorption to (GT)6-SWCNTs manifested as a nearly instantaneous quenching of the SWCNT fluorescence, leveling off to –37% integrated-fluorescence fold change (ΔF/F0) within 5 min (Figure 1b). This fluorescence quenching exhibited excellent time stability over the course of 2 h (Figure 1c). Comparing the time-dependent quenching behavior of ACE2 with (GT)6- vs (GT)15-SWCNTs affirmed the faster “leaving group” behavior of the shorter ssDNA, (GT)6, stabilizing within 10 min, as compared to (GT)15, requiring at least 60 min (Figure S3). Noncovalent ACE2 adsorption, as opposed to covalent modification, was confirmed by retention of SWCNT absorbance peaks representing various SWCNT chiralities’ electronic transitions (Figure S4). These results of decreased nIR SWCNT fluorescence emission while the absorption peaks remain unchanged demonstrate that ACE2 adsorption on SWCNTs leads to exciton quenching, involving a decreased exciton recombination frequency and/or increased nonradiative decay pathways.38
Figure 1.
Adsorption of ACE2 sensing proteins to (GT)6-SWCNTs to form nanosensor constructs. (a) Schematic depiction of ACE2-SWCNT nanosensor formation, with sensing protein ACE2. (b) ACE2-SWCNT complexation was observed as quenching of the intrinsic SWCNT near-infrared fluorescence following 1 h incubation of 6.25 mg/L ACE2 with 2.5 mg/L (GT)6-SWCNTs (final concentrations). (c) ACE2-SWCNT construct demonstrated time-stable quenched fluorescence. All fluorescence measurements were obtained with 721 nm laser excitation. Gray bars represent the standard error between experimental replicates (N = 3). (d) Adsorption of ACE2 on the SWCNT surface led to (GT)6 desorption, tracked by Cy5-labeled ssDNA following addition of 6.25 mg/L ACE2 with 2.5 mg/L Cy5-(GT)6-SWCNTs (final concentrations). The increase in Cy5-(GT)6 fluorescence from the initial quenched state on the SWCNT serves as a proxy for ACE2 adsorption. Shaded error bars represent the standard error between experimental replicates (N = 3).
The affinity of ACE2 for the ssDNA-wrapped SWCNT surface was assessed by the corona exchange assay.36 For this assay, Cy5-labeled (GT)6 ssDNA was tracked as it desorbed from the SWCNT surface and thus dequenched in the presence of ACE2. ACE2 displayed high affinity for the SWCNT surface, as ACE2 adsorption led to an 80.5% increase in Cy5 fluorescence, denoting free ssDNA, 1 h post addition of ACE2 (Figure 1d) in an ACE2 concentration-dependent manner (Figure S5). We further assessed the stability of the ACE2-SWCNT interface with a surfactant displacement assay, which confirmed strong and stable adsorption of ACE2 to the SWCNT (Figure S6).39,40 Taken together, these results suggest that ACE2 adsorbs to the SWCNT surface, displaces ssDNA originally on the SWCNT surface, and forms a stable ACE2-SWCNT conjugate that can be tested for its utility as a CoV-2 nanosensor.
NANOSENSOR RESPONSE TO THE SARS-COV-2 SPIKE PROTEIN
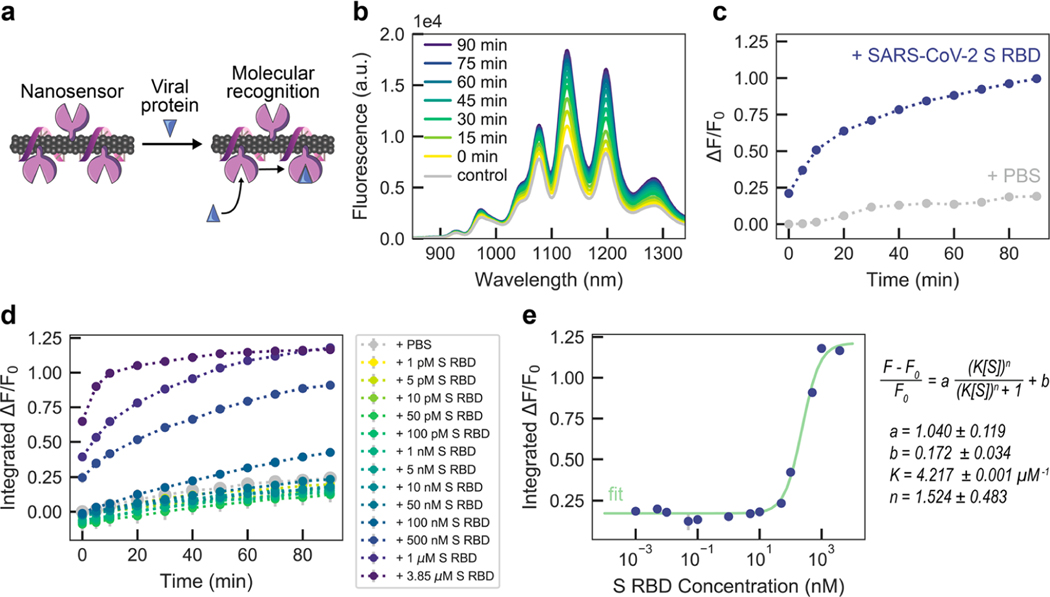
We analyzed the fluorescence response of ACE2-SWCNT nanosensors to the SARS-CoV-2 S RBD analyte, schematically represented in Figure 2a. Recognition of the CoV-2 S RBD by the nanosensor elicited a strong turn-on fluorescence response upon addition of 10 mg/L final concentration of CoV-2 S RBD to the nanosensor (formed by adsorbing 6.25 mg/L ACE2 to 2.5 mg/L (GT)6-SWCNTs) (Figure 2b). The normalized change in fluorescence of the 1130 nm SWCNT emission peak instantaneously increased to ΔF/F0 = 21.1%, reaching ΔF/F0 = 99.6% after 90 min (Figure 2c). This fluorescence modulation was verified to arise from the S RBD analyte itself rather than any impurities remaining after gel filtration chromatography (see Methods in the Supporting Information) by testing the filtrate of S RBD solution below a 3 kDa molecular weight cutoff centrifugal filter (Figure S7), which showed a negligible change in fluorescence above that of adding PBS. The concentration-dependent nanosensor response to S RBD (Figure 2d) gives rise to a 12.6 nM nanosensor LOD (see calculation in the Supporting Information). Further, approximate values for the nanosensor kinetic parameters were determined by fitting the 90 min nanosensor response to this analyte concentration series to the Hill Equation (cooperative binding model).26 Here, the integrated-fluorescence fold change of the nanosensor was correlated to the concentration of the S RBD analyte as shown in Figure 2e, resulting in an equilibrium dissociation constant (Kd) of 4.22 μM–1. These fit values represent conservative estimates for the nanosensor kinetic parameters by using the full integrated-fluorescence fold change. Moreover, this model implicates the assumption that the bulk analyte concentration remains constant (i.e., only a small fraction of total injected analyte is bound by the nanosensor). Importantly, the response of ACE2-SWCNT nanosensors for S RBD via molecular recognition was confirmed by showing insignificant (GT)6 ssDNA desorption that does not scale with injected S RBD analyte concentration (Figure S8a). Furthermore, addition of S RBD to (GT)6-SWCNTs alone (without ACE2 sensing moiety) resulted in aggregation, as the ssDNA displaced from the SWCNT surface continued to increase linearly over 6 h (Figure S8b).
Figure 2.
ACE2-SWCNT nanosensor response to SARS-CoV-2 spike protein receptor-binding domain (S RBD). (a) Schematic depiction of ACE2-SWCNT nanosensor interacting with viral protein, S RBD. Addition of 10 mg/L S RBD (final concentration) to ACE2-SWCNTs (formed by 6.25 mg/L ACE2 and 2.5 mg/L (GT)6-SWCNTs) yielded a strong turn-on fluorescence response, as shown by (b) the full fluorescence spectrum and (c) the normalized change in fluorescence (ΔF/F0) of the 1130 nm SWCNT emission peak as a function of time, over 90 min. (d) Varying S RBD concentrations were injected into ACE2-SWCNTs and the integrated-fluorescence fold change (ΔF/F0) was monitored over 90 min. (e) Integrated ΔF/F0 values at time = 90 min for varying S RBD concentrations were fit to a cooperative binding model to quantify nanosensor kinetic parameters. Fit parameters are listed with 95% confidence intervals evaluated using the t-distribution. All fluorescence measurements were obtained with 721 nm laser excitation. (c)–(e) Gray bars represent the standard error between experimental replicates (N = 3).
Nanosensor colloidal stability was verified by demonstrating that the nanosensor response to S RBD persisted after centrifugation (16.1 krcf, 30 min; Figure S9a) and overnight incubation at ambient conditions (Figure S9b). Repeating the surfactant displacement experiment with the ACE2-SWCNT nanosensor in the presence of S RBD showed that the bound receptor–ligand state further stabilized the nanosensor surface to surfactant perturbations (Figure S6e,f). This latter result, together with the retained solution stability of the analyte-bound state, indicates ACE2 was not removed from the SWCNT surface upon S RBD binding. As such, we postulate that S RBD binding to ACE2 induces a conformational change pinning ACE2 to the SWCNT surface, thus simultaneously increasing SWCNT fluorescence emission via exciton dequenching and stabilizing the SWCNT surface against surfactant interaction.
NANOSENSOR ANALYTE SELECTIVITY AND BIOENVIRONMENT ROBUSTNESS
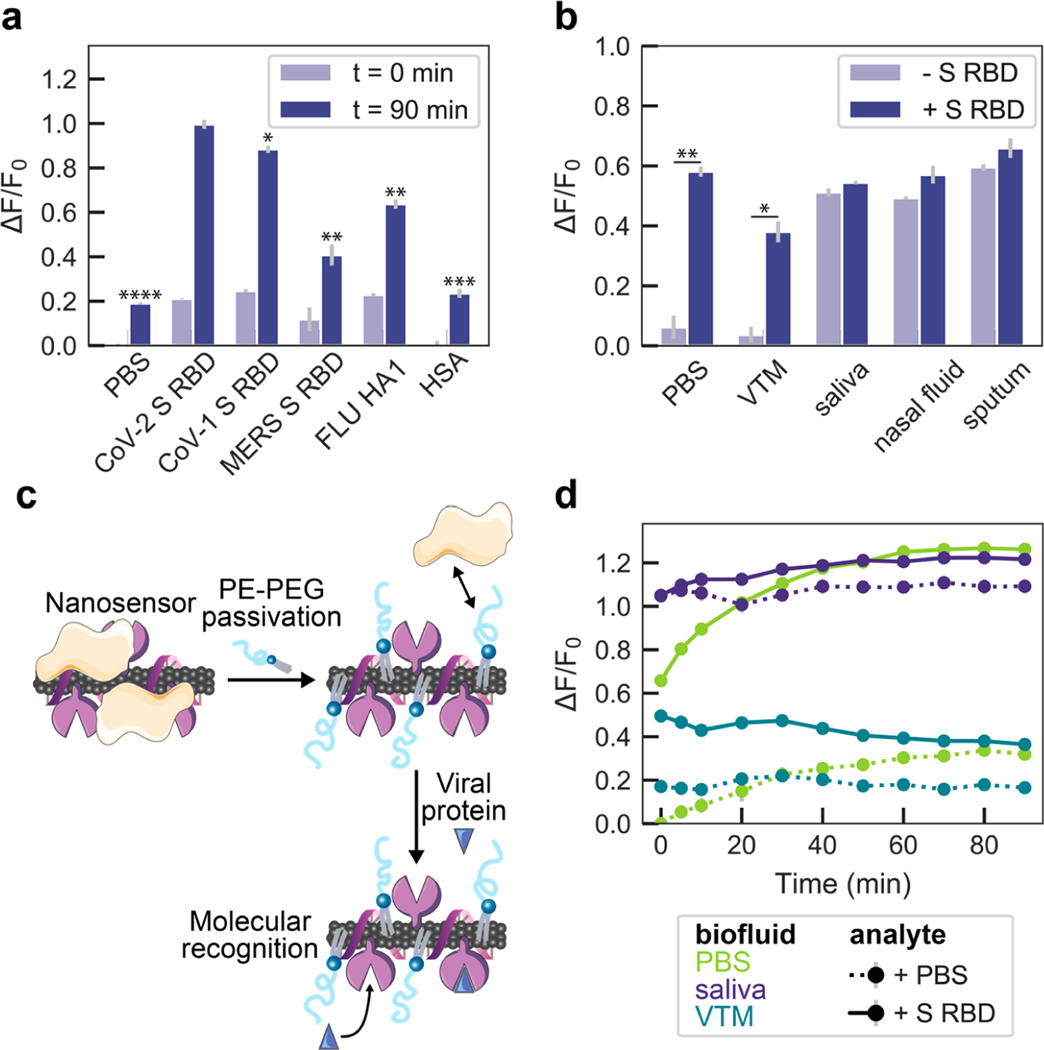
We next investigated the selectivity of ACE2-SWCNT nanosensors to a panel of viral spike-like proteins. This viral analyte panel was composed of the SARS-CoV-2 S RBD in addition to the SARS-CoV-1 S RBD, MERS S RBD, and FLU hemagglutinin subunit (HA1). Serum albumin (HSA) was also included as a protein abundant in bioenvironments and in viral transport medium (VTM; 2% Fetal Bovine Serum, 100 μg/mL Gentamicin, 0.5 μg/mL Amphotericin in a Hanks Balanced salt solution base). Viral proteins were normalized on a mass basis (10 mg/L final concentration) to account for varying molecular weights. SARS-CoV-2 S RBD elicited the largest nanosensor response of ΔF/F0 = 99.6% at the 1130 nm SWCNT emission peak after 90 min (Figure 3a), followed by SARS-CoV-1 S RBD (ΔF/F0 = 88.4%). This cross-reactivity is expected, as ACE2 is also the cell membrane protein that binds to CoV-1 S RBD, although at ∼10–20-fold lower affinity.41,42 MERS and FLU spike-like proteins naturally interact with different cell membrane receptors, accounting for this lower magnitude fluorescence response with our ACE2-SWCNT nanosensors.
Figure 3.
ACE2-SWCNT nanosensor selectivity and sensitivity in biofluid environments. (a) Normalized change in fluorescence (ΔF/F0) of the 1130 nm SWCNT emission peak for the ACE2-SWCNT nanosensor 0 and 90 min after exposure to 10 mg/L of viral protein panel: SARS-CoV-2 spike receptor-binding domain (S RBD), SARS-CoV-1 S RBD, MERS S RBD, and FLU hemagglutinin subunit (HA1). ****P = 0.0006 (PBS), ***P = 0.0014 (HSA), **P = 0.0065 (FLU) and 0.0076 (MERS), and *P = 0.0503 (SARS-CoV-1) in independent two-sample t tests, for each analyte ΔF/F0 response at t = 90 min in comparison to SARS-CoV-2 S RBD. (b) ACE2-SWCNT nanosensor response 90 min after exposure to 1 μM S RBD in the presence of 1% relevant biofluids: viral transport medium (VTM), saliva, nasal fluid, and sputum (treated with sputasol). **P = 0.0065 (PBS) and *P = 0.0161 (VTM) in independent two-sample t tests, for ΔF/F0 response in biofluids compared before vs after S RBD addition. (c) Schematic depiction of nanosensor biofouling with proteins present in relevant biofluids, mitigated upon passivation with phosphatidylethanolamine phospholipid with a 5000 Da PEG chain (PE–PEG). (d) Response of PE–PEG passivated nanosensor to 500 nM S RBD in the presence of PBS, 10% VTM, or 1% saliva. Surface passivation with a hydrophilic polymer improved the nanosensor response that was otherwise greatly attenuated, as shown in (b). All fluorescence measurements were obtained with 721 nm laser excitation. Gray bars represent the standard error between experimental replicates (N = 3).
Nanosensor compatibility in biofluids was assessed by testing the nanosensor response to CoV-2 S RBD in 1% relevant biological fluids, including viral transport medium (VTM), human saliva, human nasal fluid, and human sputum (treated with sputasol) (biofluid details in Table S1). Although the nanosensor response was maintained in PBS and VTM, the response was diminished in the other biofluids, with a ΔF/F0 = 3.2% in saliva, 7.8% in nasal fluid, and 6.3% in sputum (Figure 3b). The attenuation of nanosensor response seems to arise from biofluid protein adsorption to the nanosensor surface that raises the baseline fluorescence and obfuscates viral analyte interaction, whereby the nanosensor fluorescence in the biofluids alone is stable with an increased baseline fluorescence (Figure S9c).
To mitigate the unfavorable effects of biofouling that lead to this diminished nanosensor response, we pursued a passivation strategy involving phosphatidylethanolamine phospholipid with a 5000 Da PEG chain attached to the headgroup (PE–PEG), schematically represented in Figure 3c.43 The PE–PEG passivated nanosensor response to 500 nM CoV-2 S RBD was ΔF/F0 = 19.9% in 10% VTM (otherwise absent without passivation) and ΔF/F0 = 12.4% in 1% saliva (otherwise 3.2% without passivation), suggesting PE–PEG nanosensor passivation enables partial reduction of nanosensor biofouling.
IMMOBILIZED NANOSENSOR RESPONSE TO THE SARS-COV-2 SPIKE PROTEIN AND VIRUS-LIKE PARTICLES
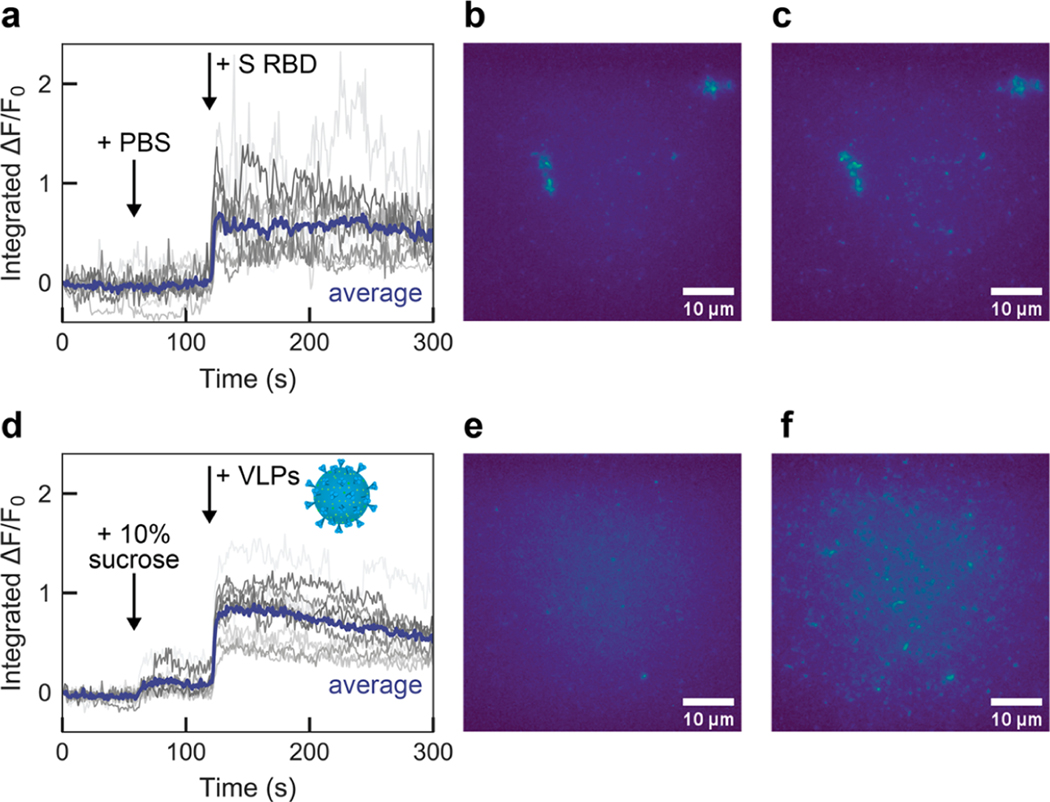
We then translated the nanosensors from in-solution sensing to a surface-immobilized format for imaging. ACE2-SWCNTs (formed by adsorbing 12.5 mg/L ACE2 to 5 mg/L (GT)6-SWCNTs) were immobilized on a glass-bottom microwell dish and imaged with a 100× oil immersion objective (Figure 4). Addition of PBS at 60 s did not cause a change in fluorescence signal, as anticipated from solution-based nanosensor control experiments (Figure 2). Upon injection of 2 μM (final concentration) S RBD, the average integrated-fluorescence intensity change was ΔF/F0 = 65.1% within 5 s (Figure 4a–c). This experiment was repeated using virus-like particles (VLPs), which are formed by coexpressing all four SARS-CoV-2 structural proteins (spike, membrane, nucleocapsid, and envelope proteins). Addition of 10% sucrose (the VLP buffer) at 60 s slightly increased the baseline fluorescence. Injection of 35 mg/L VLPs increased the average integrated-fluorescence intensity by ΔF/F0 = 72.8% within 5 s (Figure 4d–f). This concentration of VLPs corresponds to approximately 17 nM S RBD. To evaluate the specificity of the observed nanosensor response, we then tested VLPs produced with and without S protein coexpressed. We found that the immobilized nanosensor exhibited a response of ΔF/F0 = 19.4% within 5 s for the VLPs without S protein, compared to a response of ΔF/F0= 70.7% for the VLPs with S protein (Figure S10).
Figure 4.
Surface-immobilized ACE2-SWCNT nanosensor response to SARS-CoV-2 spike protein receptor-binding domain (S RBD) and virus-like particles (VLPs). Microscopy traces of ACE2-SWCNTs (formed by 12.5 mg/L ACE2 and 5 mg/L (GT)6-SWCNTs) immobilized on a glass-bottom microwell dish exhibited a fluorescence response to both S RBD and VLPs, for single regions of interest (gray; 12 total per image) and the average intensity (purple). (a)–(c) Addition of PBS at 60 s caused no change in fluorescence, as expected, and addition of 2 μM S RBD (final concentration) at 120 s yielded a turn-on fluorescence response, as shown by (a) the integrated-fluorescence fold change (ΔF/F0) over 5 min and entire field-of-view at (b) time = 0 s and (c) time = 125 s. (d)–(f) Addition of 10% sucrose buffer (to match VLP buffer) at 60 s caused a slight increase in fluorescence and addition of 35 mg/L VLPs (final concentration) at 120 s yielded a turn-on fluorescence response, as shown by (d) the integrated-fluorescence fold change (ΔF/F0) over 5 min and entire field-of-view at (e) time = 0 s and (f) time = 125 s. All fluorescence images were obtained with 721 nm laser excitation and a 100× oil immersion objective. VLP adapted with permission from an image by Maya Peters Kostman for the Innovative Genomics Institute.
CONCLUSION
In summary, we have developed an optical SWCNT-based nanosensor capable of detecting SARS-CoV-2 via S protein recognition. Our protein-SWCNT design concept can be extended to incorporate other proteins of interest and for applications beyond biological sensing, such as the label-free study of protein–protein interactions. To construct these nanosensors, ACE2 was noncovalently adsorbed to the SWCNT surface and, in the presence of the viral S protein analyte, binding elicited a modulation of the intrinsic SWCNT fluorescence. We studied ACE2 adsorption to SWCNTs by employing corona exchange and surfactant displacement assays, which confirmed stable adsorption of ACE2 to SWCNTs, and subsequently confirmed S RBD binding to the ACE2-SWCNT nanosensor. The resulting nanosensors displayed excellent colloidal stability and retained binding capability to CoV-2 S RBD when surface-immobilized. Nanosensors exhibited a 100% turn-on response in fluorescence upon addition of 1 μM CoV-2 S RBD, with response scaling as a function of concentration. Fitting to a cooperative binding model gave rise to kinetic parameter estimates to quantify nanosensor performance. Although the solution-phase LOD (12.6 nM S RBD) remains above that of realistic samples (viral loads ranging from ∼101–104 viral copies per μL,44–47 translating to ∼0.005–5 pM S RBD), the surface-immobilized nanosensor can achieve a ΔF/F0 = 73% within 5 s of exposure to 35 mg/L VLPs as a mimic of the full SARS-CoV-2 virion, with detection able to reach down to ∼104–106 viral copies per μL (Figure S11). The lower magnitude response for VLPs without S protein (ΔF/F0 = 19%) supports our hypothesis that molecular recognition is enabled by the specific ACE2-S protein interaction. Furthermore, solution-phase nanosensor passivation with a hydrophilic polymer (PEG) attached to phospholipids (PE) provided some improvement of nanosensor response to S protein in 10% VTM and 1% saliva. However, additional strategies must be pursued to further abate biofouling while retaining the fluorescence response, thus increasing the feasibility of these nanosensors to function in crude biofluids. Such strategies may include varying the antibiofouling polymers attached to the phospholipids, such as incorporating zwitterionic polymers,35,48 or covalently linking polymers directly to the SWCNT surface.49,50 Future work will also improve nanosensor sensitivity and selectivity to the CoV-2 S protein necessary for clinical application (comparisons in Table S2), potentially by incorporating a more specific sensing protein than ACE2, which serves as a receptor for other viral and endogenous proteins.51 Such sensing proteins could include antibodies52 or nanobodies,53,54 as well as other viral biomarker binding moieties. Moreover, various protein-anchoring55,56 and SWCNT precoating43,57 strategies can be pursued to give rise to a more prequenched initial nanosensor fluorescence, thus leading to a larger magnitude increase in the presence of the viral analyte. Incorporating nanosensors in form factors such as paper or hydrogel may enable locally concentrating viral analytes to elicit a greater fluorescence response, or immobilizing nanosensors in a flow channel may allow an accumulation mode of function. Finally, patient biofluids may undergo a facile filtering step to concentrate proteins.58
Early and frequent testing is key to trace and control the spread of COVID-19. However, current diagnostics suffer from insufficient supply and throughput, where our reliance on tests with long turnaround times leads to delays in patients receiving test results. A technology capable of rapidly detecting active infections in crude biofluids is needed. Taken together, our data show that SWCNT-based nanosensors noncovalently functionalized with the human ACE2 receptor can detect the SARS-CoV-2 S protein and can be immobilized and imaged on microfluidic surfaces. Though less sensitive than PCR-based testing, the rapid nanosensor response in the surface-immobilized state toward SARS-CoV-2 VLPs has distinct advantages in enabling on-site testing and has the potential to detect SARS-CoV-2 without patient biofluid sample processing and purification. Ultimately, these nanosensors can be incorporated into a point-of-care device for rapid diagnosis of individuals actively infected with SARS-CoV-2, using accessible equipment from a different supply chain than that of current testing modes.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support of the Innovative Genomics Institute LGR ERA, Citris/Banatao Seed Funding, and Facebook. We acknowledge support of an NIH NIDA CEBRA Award # R21DA044010 (to M.P.L.), a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI) (to M.P.L.), the Simons Foundation (to M.P.L.), a Stanley Fahn PDF Junior Faculty Grant with Award # PF-JFA-1760 (to M.P.L.), a Beckman Foundation Young Investigator Award (to M.P.L.), and a DARPA Young Investigator Award (to M.P.L.). M.P.L. is a Chan Zuckerberg Biohub investigator. R.L.P., F.L., and D.Y. acknowledge the support of NSF Graduate Research Fellowships (NSF DGE 1752814). R.L.P. and M.P.L. designed the research. R.L.P. performed the research. R.L.P., F.L., D.Y., N.N., S.J., and M.P.L. assisted in experimental optimization and methods. J.E.P., L.K., Y.C.C., Y.W.C., and H.Y.S. optimized SARS-CoV-2 protein material production and provided material and expertise. R.L.P. analyzed the data. All authors discussed the research and results. R.L.P., F.L., and M.P.L. wrote the paper.
ABBREVIATIONS
- ACE2
angiotensin-converting enzyme 2
- GT
guanine-thymine
- HA1
hemagglutinin subunit
- HSA
human serum albumin
- LOD
limit of detection
- MERS
Middle East respiratory syndrome
- nIR
near-infrared
- PBS
phosphate-buffered saline
- PE–PEG
phosphatidylethanolamine-poly-ethylene glycol
- RT-PCR
reverse transcription polymerase chain reaction
- SARS-CoV
severe acute respiratory syndrome coronavirus
- ssDNA
single-stranded DNA
- SWCNT
single-walled carbon nanotube
- SC
sodium cholate
- S RBD
spike protein receptor-binding domain
- VTM
viral transport medium
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c00118.
Methods, extended discussion on results; fluorescence spectra, absorbance spectra, nanosensor response (Figures S1–S11); purchased biofluid and protein specifications (Table S1), comparison of sensor performance to current SARS-CoV-2 diagnostic technologies (Table S2) (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.1c00118
The authors declare no competing financial interest.
Contributor Information
Rebecca L. Pinals, Department of Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States
Francis Ledesma, Department of Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States.
Darwin Yang, Department of Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States.
Nicole Navarro, Department of Chemistry, University of California, Berkeley, California 94720, United States.
Sanghwa Jeong, Department of Chemical and Biomolecular Engineering, University of California, Berkeley, California 94720, United States.
John E. Pak, Chan Zuckerberg Biohub, San Francisco, California 94158, United States
Lili Kuo, Wadsworth Center, New York State Department of Health, Slingerlands, New York 12159, United States.
Yung-Chun Chuang, Leadgene Biomedical Inc., Tainan 71042, Taiwan; Department of Medical Laboratory Science and Biotechnology, College of Medicine, National Cheng Kung University, Tainan 70101, Taiwan.
Yu-Wei Cheng, Leadgene Biomedical Inc., Tainan 71042, Taiwan.
Hung-Yu Sun, Institute of Molecular Medicine, College of Medicine, National Cheng Kung University, Tainan 70101, Taiwan.
Markita P. Landry, Department of Chemical and Biomolecular Engineering and California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, California 94720, United States; Chan Zuckerberg Biohub, San Francisco, California 94158, United States; Innovative Genomics Institute (IGI), Berkeley, California 94720, United States
REFERENCES
- (1).WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int (accessed Feb 15, 2021).
- (2).Poletti P; Tirani M; Cereda D; Trentini F; Guzzetta G; Sabatino G; Marziano V; Castrofino A; Grosso F; Del Castillo G; Piccarreta R; ATS Lombardy COVID-19 Task Force; Andreassi A; Melegaro A; Gramegna M; Ajelli M; Merler S. Probability of Symptoms and Critical Disease after SARS-CoV-2 Infection. arXiv:2006.08471; https://arxiv.org/abs/2006.08471v2 (accessed October 6, 2020). [Google Scholar]
- (3).Li Q; Guan X; Wu P; Wang X; Zhou L; Tong Y; Ren R; Leung KSM; Lau EHY; Wong JY; Xing X; Xiang N; Wu Y; Li C; Chen Q; Li D; Liu T; Zhao J; Liu M; Tu W; Chen C; Jin L; Yang R; Wang Q; Zhou S; Wang R; Liu H; Luo Y; Liu Y; Shao G; Li H; Tao Z; Yang Y; Deng Z; Liu B; Ma Z; Zhang Y; Shi G; Lam TTY; Wu JT; Gao GF; Cowling BJ; Yang B; Leung GM; Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med 2020, 382 (13), 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Esbin MN; Whitney ON; Chong S; Maurer A; Darzacq X; Tjian R. Overcoming the Bottleneck to Widespread Testing: A Rapid Review of Nucleic Acid Testing Approaches for COVID-19 Detection. RNA 2020, 26 (7), 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mio C; Cifu A; Marzinotto S; Bergamin N; Caldana C; Cattarossi S; Cmet S; Cussigh A; Martinella R; Zucco J; Verardo R; Schneider C; Marcon B; Zampieri S; Pipan C; Curcio F. A Streamlined Approach to Rapidly Detect SARS-CoV-2 Infection Avoiding RNA Extraction: Workflow Validation. Dis. Markers 2020, 2020, 8869424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sentmanat M; Kouranova E; Cui X. One-Step RNA Extraction for RT-QPCR Detection of 2019-NCoV. Bio-Rxiv:2020.04.02.022384 DOI: 10.1101/2020.04.02.022384 (accessed October 6, 2020). [DOI] [Google Scholar]
- (7).Won J; Lee S; Park M; Kim TY; Park MG; Choi BY; Kim D; Chang H; Kim VN; Lee CJ Development of a Laboratory-Safe and Low-Cost Detection Protocol for SARS-CoV-2 of the Coronavirus Disease 2019 (COVID-19). Experimental Neurobiology 2020, 29 (2), 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).McMinn S; Carlsen A; Jaspers B; Talbot R; Adeline S. In Large Texas Cities, Access To Coronavirus Testing May Depend On Where You Live. Shots: Health News from NPR; NPR, May 27, 2020;https://www.npr.org/sections/health-shots/2020/05/27/862215848/across-texas-black-and-hispanic-neighborhoods-have-fewer-coronavirus-testing-sit (accessed October 6, 2020). [Google Scholar]
- (9).Collins K. Is Your State Doing Enough Coronavirus Testing? The New York Times; New York Times Company, November 1, 2020; https://www.nytimes.com/interactive/2020/us/coronavirus-testing.html (accessed February 17, 2021). [Google Scholar]
- (10).Yu L; Wu S; Hao X; Dong X; Mao L; Pelechano V; Chen W-H; Yin X. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin. Chem 2020, 66 (7), 975–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Qiu G; Gai Z; Tao Y; Schmitt J; Kullak-Ublick GA; Wang J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14 (5), 5268–5277. [DOI] [PubMed] [Google Scholar]
- (12).Broughton JP; Deng X; Yu G; Fasching CL; Servellita V; Singh J; Miao X; Streithorst JA; Granados A; Sotomayor-Gonzalez A; Zorn K; Gopez A; Hsu E; Gu W; Miller S; Pan C-Y; Guevara H; Wadford DA; Chen JS; Chiu CY CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol 2020, 38 (7), 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Guglielmi G. The Explosion of New Coronavirus Tests That Could Help to End the Pandemic. Nature 2020, 583 (7817), 506–509. [DOI] [PubMed] [Google Scholar]
- (14).Guglielmi G. Fast Coronavirus Tests: What They Can and Can’t Do. Nature 2020, 585 (7826), 496–498. [DOI] [PubMed] [Google Scholar]
- (15).Du L; He Y; Zhou Y; Liu S; Zheng B-J; Jiang S. The Spike Protein of SARS-CoV — a Target for Vaccine and Therapeutic Development. Nat. Rev. Microbiol 2009, 7 (3), 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lan J; Ge J; Yu J; Shan S; Zhou H; Fan S; Zhang Q; Shi X; Wang Q; Zhang L; Wang X. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215. [DOI] [PubMed] [Google Scholar]
- (17).Sui J; Li W; Murakami A; Tamin A; Matthews LJ; Wong SK; Moore MJ; Tallarico ASC; Olurinde M; Choe H; Anderson LJ; Bellini WJ; Farzan M; Marasco WA Potent Neutralization of Severe Acute Respiratory Syndrome (SARS) Coronavirus by a Human MAb to S1 Protein That Blocks Receptor Association. Proc. Natl. Acad. Sci. U. S. A 2004, 101 (8), 2536–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bar-On YM; Flamholz A; Phillips R; Milo R. SARS-CoV-2 (COVID-19) by the Numbers. eLife 2020, 9, No. e57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Seo G; Lee G; Kim MJ; Baek S-H; Choi M; Ku KB; Lee C-S; Jun S; Park D; Kim HG; Kim S-J; Lee J-O; Kim BT; Park EC; Kim SI Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14 (4), 5135–5142. [DOI] [PubMed] [Google Scholar]
- (20).Zhu X; Wang X; Han L; Chen T; Wang L; Li H; Li S; He L; Fu X; Chen S; Xing M; Chen H; Wang Y. Multiplex Reverse Transcription Loop-Mediated Isothermal Amplification Combined with Nanoparticle-Based Lateral Flow Biosensor for the Diagnosis of COVID-19. Biosens. Bioelectron 2020, 166, 112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Udugama B; Kadhiresan P; Kozlowski HN; Malekjahani A; Osborne M; Li VYC; Chen H; Mubareka S; Gubbay JB; Chan WCW Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14 (4), 3822–3835. [DOI] [PubMed] [Google Scholar]
- (22).Moitra P; Alafeef M; Dighe K; Frieman MB; Pan D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14 (6), 7617–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Teengam P; Siangproh W; Tuantranont A; Vilaivan T; Chailapakul O; Henry CS Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem 2017, 89 (10), 5428–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang J; Landry MP; Barone PW; Kim J-H; Lin S; Ulissi ZW; Lin D; Mu B; Boghossian AA; Hilmer AJ; Rwei A; Hinckley AC; Kruss S; Shandell MA; Nair N; Blake S; S̨en F; S̨en S; Croy RG; Li D; Yum K; Ahn J-H; Jin H; Heller DA; Essigmann JM; Blankschtein D; Strano MS Molecular Recognition Using Corona Phase Complexes Made of Synthetic Polymers Adsorbed on Carbon Nanotubes. Nat. Nanotechnol 2013, 8 (12), 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Boghossian AA; Zhang J; Barone PW; Reuel NF; Kim J-H; Heller DA; Ahn J-H; Hilmer AJ; Rwei A; Arkalgud JR; Zhang CT; Strano MS Near-Infrared Fluorescent Sensors Based on Single-Walled Carbon Nanotubes for Life Sciences Applications. ChemSusChem 2011, 4 (7), 848–863. [DOI] [PubMed] [Google Scholar]
- (26).Kruss S; Landry MP; Vander Ende E; Lima BMA; Reuel NF; Zhang J; Nelson J; Mu B; Hilmer A; Strano M. Neurotransmitter Detection Using Corona Phase Molecular Recognition on Fluorescent Single-Walled Carbon Nanotube Sensors. J. Am. Chem. Soc 2014, 136 (2), 713–724. [DOI] [PubMed] [Google Scholar]
- (27).Salem DP; Gong X; Liu AT; Akombi K; Strano MS Immobilization and Function of NIR-Fluorescent Carbon Nanotube Sensors on Paper Substrates for Fluidic Manipulation. Anal. Chem 2020, 92 (1), 916–923. [DOI] [PubMed] [Google Scholar]
- (28).Kozawa D; Cho S-Y; Gong X; Nguyen FT; Jin X; Lee MA; Lee H; Zeng A; Xue G; Schacherl J; Gibson S; Vega L; Strano MS A Fiber Optic Interface Coupled to Nanosensors: Applications to Protein Aggregation and Organic Molecule Quantification. ACS Nano 2020, 14 (8), 10141–10152. [DOI] [PubMed] [Google Scholar]
- (29).Wong MH; Giraldo JP; Kwak S-Y; Koman VB; Sinclair R; Lew TTS; Bisker G; Liu P; Strano MS Nitroaromatic Detection and Infrared Communication from Wild-Type Plants Using Plant Nanobionics. Nat. Mater 2017, 16 (2), 264–272. [DOI] [PubMed] [Google Scholar]
- (30).Antonucci A; Kupis-Rozmysłowicz J; Boghossian AA Noncovalent Protein and Peptide Functionalization of Single-Walled Carbon Nanotubes for Biodelivery and Optical Sensing Applications. ACS Appl. Mater. Interfaces 2017, 9 (13), 11321–11331. [DOI] [PubMed] [Google Scholar]
- (31).Zubkovs V; Schuergers N; Lambert B; Ahunbay E; Boghossian AA Mediatorless, Reversible Optical Nanosensor Enabled through Enzymatic Pocket Doping. Small 2017, 13 (42), 1701654. [DOI] [PubMed] [Google Scholar]
- (32).Gillen AJ; Boghossian AA Non-Covalent Methods of Engineering Optical Sensors Based on Single-Walled Carbon Nanotubes. Front. Chem 2019, 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Nelson JT; Kim S; Reuel NF; Salem DP; Bisker G; Landry MP; Kruss S; Barone PW; Kwak S; Strano MS Mechanism of Immobilized Protein A Binding to Immunoglobulin G on Nanosensor Array Surfaces. Anal. Chem 2015, 87 (16), 8186–8193. [DOI] [PubMed] [Google Scholar]
- (34).Salem DP; Gong X; Lee H; Zeng A; Xue G; Schacherl J; Gibson S; Strano MS Characterization of Protein Aggregation Using Hydrogel-Encapsulated NIR Fluorescent Nanoparticle Sensors. ACS Sens. 2020, 5 (2), 327–337. [DOI] [PubMed] [Google Scholar]
- (35).Pinals RL; Chio L; Ledesma F; Landry MP Engineering at the Nano-Bio Interface: Harnessing the Protein Corona towards Nanoparticle Design and Function. Analyst 2020, 145 (15), 5090–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pinals RL; Yang D; Lui A; Cao W; Landry MP Corona Exchange Dynamics on Carbon Nanotubes by Multiplexed Fluorescence Monitoring. J. Am. Chem. Soc 2020, 142 (3), 1254–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Pinals RL; Yang D; Rosenberg DJ; Chaudhary T; Crothers AR; Iavarone AT; Hammel M; Landry MP Quantitative Protein Corona Composition and Dynamics on Carbon Nanotubes in Biological Environments. Angew. Chem., Int. Ed 2020, 59 (52), 23668–23677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Heller DA; Pratt GW; Zhang J; Nair N; Hansborough AJ; Boghossian AA; Reuel NF; Barone PW; Strano MS Peptide Secondary Structure Modulates Single-Walled Carbon Nanotube Fluorescence as a Chaperone Sensor for Nitroaromatics. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (21), 8544–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Alizadehmojarad AA; Zhou X; Beyene AG; Chacon KE; Sung Y; Pinals RL; Landry MP; Vukovic,´L. Binding Affinity and Conformational Preferences Influence Kinetic Stability of Short Oligonucleotides on Carbon Nanotubes. Adv. Mater. Interfaces 2020, 7 (15), 2000353. [Google Scholar]
- (40).Beyene AG; Alizadehmojarad AA; Dorlhiac G; Goh N; Streets AM; Král P; Vuković L; Landry MP Ultralarge Modulation of Fluorescence by Neuromodulators in Carbon Nanotubes Functionalized with Self-Assembled Oligonucleotide Rings. Nano Lett. 2018, 18 (11), 6995–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Tai W; He L; Zhang X; Pu J; Voronin D; Jiang S; Zhou Y; Du L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol 2020, 17 (6), 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wrapp D; Wang N; Corbett KS; Goldsmith JA; Hsieh C-L; Abiona O; Graham BS; McLellan JS Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367 (6483), 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Yang D; Yang SJ; Del Bonis-O’Donnell JT; Pinals RL; Landry MP Mitigation of Carbon Nanotube Neurosensor Induced Transcriptomic and Morphological Changes in Mouse Microglia with Surface Passivation. ACS Nano 2020, 14 (10), 13794–13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zou L; Ruan F; Huang M; Liang L; Huang H; Hong Z; Yu J; Kang M; Song Y; Xia J; Guo Q; Song T; He J; Yen H-L; Peiris M; Wu J. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med 2020, 382 (12), 1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Pan Y; Zhang D; Yang P; Poon LLM; Wang Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis 2020, 20 (4), 411–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).To KK-W; Tsang OT-Y; Leung W-S; Tam AR; Wu T-C; Lung DC; Yip CC-Y; Cai J-P; Chan JM-C; Chik TS-H; Lau DP-L; Choi CY-C; Chen L-L; Chan W-M; Chan K-H; Ip JD; Ng AC-K; Poon RW-S; Luo C-T; Cheng VC-C; Chan JF-W; Hung IF-N; Chen Z; Chen H; Yuen K-Y Temporal Profiles of Viral Load in Posterior Oropharyngeal Saliva Samples and Serum Antibody Responses during Infection by SARS-CoV-2: An Observational Cohort Study. Lancet Infect. Dis 2020, 20 (5), 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Wyllie AL; Fournier J; Casanovas-Massana A; Campbell M; Tokuyama M; Vijayakumar P; Warren JL; Geng B; Muenker MC; Moore AJ; Vogels CBF; Petrone ME; Ott IM; Lu P; Venkataraman A; Lu-Culligan A; Klein J; Earnest R; Simonov M; Datta R; Handoko R; Naushad N; Sewanan LR; Valdez J; White EB; Lapidus S; Kalinich CC; Jiang X; Kim DJ; Kudo E; Linehan M; Mao T; Moriyama M; Oh JE; Park A; Silva J; Song E; Takahashi T; Taura M; Weizman O-E; Wong P; Yang Y; Bermejo S; Odio CD; Omer SB; Dela Cruz CS; Farhadian S; Martinello RA; Iwasaki A; Grubaugh ND; Ko AI Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med 2020, 383 (13), 1283–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Shao Q; Jiang S. Molecular Understanding and Design of Zwitterionic Materials. Adv. Mater 2015, 27 (1), 15–26. [DOI] [PubMed] [Google Scholar]
- (49).Chio L; Pinals RL; Murali A; Goh NS; Landry MP Covalent Surface Modification Effects on Single-Walled Carbon Nanotubes for Targeted Sensing and Optical Imaging. Adv. Funct. Mater 2020, 30 (17), 1910556. [Google Scholar]
- (50).Kwon H; Furmanchuk A; Kim M; Meany B; Guo Y; Schatz GC; Wang Y. Molecularly Tunable Fluorescent Quantum Defects. J. Am. Chem. Soc 2016, 138 (21), 6878–6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol 2016, 3 (1), 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Lv H; Wu NC; Tsang OT-Y; Yuan M; Perera RAPM; Leung WS; So RTY; Chan JMC; Yip GK; Chik TSH; Wang Y; Choi CYC; Lin Y; Ng WW; Zhao J; Poon LLM; Peiris JSM; Wilson IA; Mok CKP Cross-Reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020, 31 (9), 107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Schoof M; Faust B; Saunders RA; Sangwan S; Rezelj V; Hoppe N; Boone M; Billesbølle CB; Puchades C; Azumaya CM; Kratochvil HT; Zimanyi M; Deshpande I; Liang J; Dickinson S; Nguyen HC; Chio CM; Merz GE; Thompson MC; Diwanji D; Schaefer K; Anand AA; Dobzinski N; Zha BS; Simoneau CR; Leon K; White KM; Chio US; Gupta M; Jin M; Li F; Liu Y; Zhang K; Bulkley D; Sun M; Smith AM; Rizo AN; Moss F; Brilot AF; Pourmal S; Trenker R; Pospiech T; Gupta S; Barsi-Rhyne B; Belyy V; Barile-Hill AW; Nock S; Liu Y; Krogan NJ; Ralston CY; Swaney DL; García-Sastre A; Ott M; Vignuzzi M; Consortium4‡, Q. S. B.; Walter P, Manglik A. An Ultrapotent Synthetic Nanobody Neutralizes SARS-CoV-2 by Stabilizing Inactive Spike. Science 2020, 370 (6523), 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Wrapp D; De Vlieger D; Corbett KS; Torres GM; Wang N; Van Breedam W; Roose K; van Schie L; Hoffmann M; Pöhlmann S; Graham BS; Callewaert N; Schepens B; Saelens X; McLellan JS Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181 (5), 1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wang S; Humphreys ES; Chung S-Y; Delduco DF; Lustig SR; Wang H; Parker KN; Rizzo NW; Subramoney S; Chiang Y-M; Jagota A. Peptides with Selective Affinity for Carbon Nanotubes. Nat. Mater 2003, 2 (3), 196–200. [DOI] [PubMed] [Google Scholar]
- (56).Dunakey SJG; Coyle BL; Thomas A; Xu M; Swift BJF; Baneyx F. Selective Labeling and Decoration of the Ends and Sidewalls of Single-Walled Carbon Nanotubes Using Mono- and Bispecific Solid-Binding Fluorescent Proteins. Bioconjugate Chem. 2019, 30 (3), 959–965. [DOI] [PubMed] [Google Scholar]
- (57).Satishkumar BC; Brown LO; Gao Y; Wang C-C; Wang H-L; Doorn SK Reversible Fluorescence Quenching in Carbon Nanotubes for Biomolecular Sensing. Nat. Nanotechnol 2007, 2 (9), 560–564. [DOI] [PubMed] [Google Scholar]
- (58).Mohamed R; Campbell J-L; Cooper-White J; Dimeski G; Punyadeera C. The Impact of Saliva Collection and Processing Methods on CRP, IgE, and Myoglobin Immunoassays. Clin Trans Med. 2012, 1 (1), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.