-
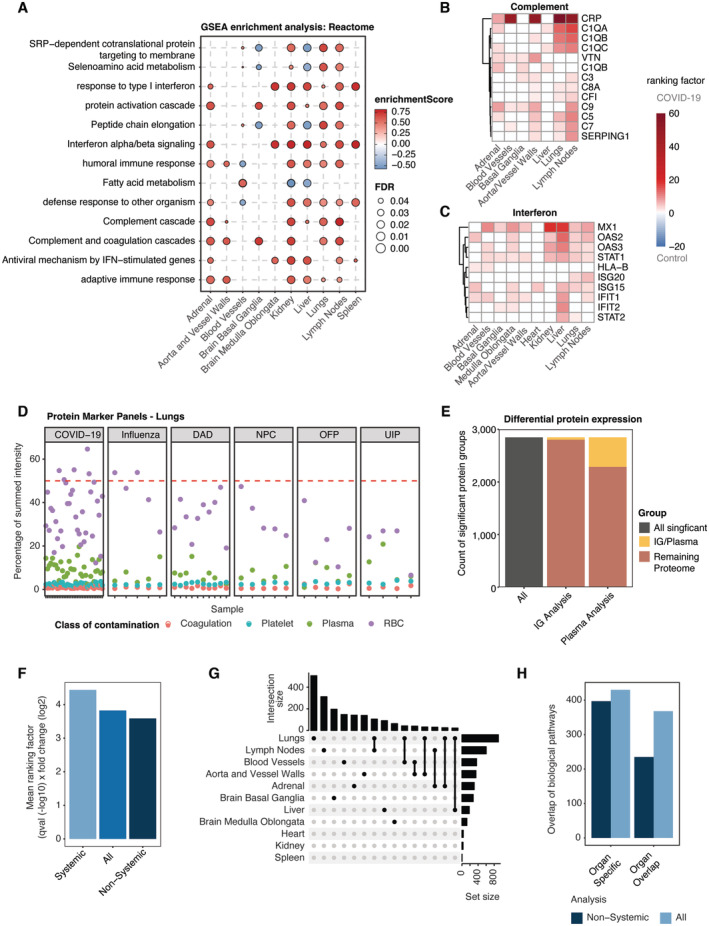
A
Terms of biological pathway enrichment analysis (GSEA) of the differential protein expression between COVID‐19 and controls and a minimal overlap of six organs in this study using the pathway Reactome database. Color codes of each enrichment term depict the enrichment score, whereas point size reflects the respective FDR.
-
B, C
Representative protein panels for the biological pathway “complement system” (B) the “interferon signaling” (C) with significance (fc > 1.5, q‐val < 0.05) in ≥ 3 organs and 2 organs, respectively. Each heat map depicts the ranking factor (q‐value (−log10) × fold change (log2)) for a protein in different organs as a measure of comparable significance.
-
D
Assessment of the prevalence for proteins in previously identified quality marker panels on coagulation, plasma, platelets and red blood cells (RBC), exemplarily shown for lung tissue. The percentage of summed protein intensities for each panel is illustrated for each sample across all disease pathologies included in our study. The ratio of 50% signal of the total protein abundance is highlighted (dashed red line).
-
E
Ratio of plasma or immunoglobulin‐related proteins on the total of significantly differentially regulated proteins (fc > 1.5, q‐val < 0.05) between COVID‐19 and control samples across all organs.
-
F
Calculation of the mean ranking factor (q‐value (−log10) × fold change (log2)) for significantly dysregulated (COVID‐19 vs. control) proteins of the original proteome (All) as well as proteins associated with the systemic and nonsystemic effects in tissue across all organs.
-
G
Upset plot depicting the intersection of significantly dysregulated (COVID‐19 vs. control) proteins across all organs.
-
H
Comparison of a biological pathway enrichment for significantly dysregulated (COVID‐19 vs. control) proteins of the original proteome and after identification of the systemic effects using the Reactome, KEGG and GO biological process databases across all organs. For each enrichment, the number of pathways is shown for organ specificity and overlap in at least two organs.