FIGURE 4.
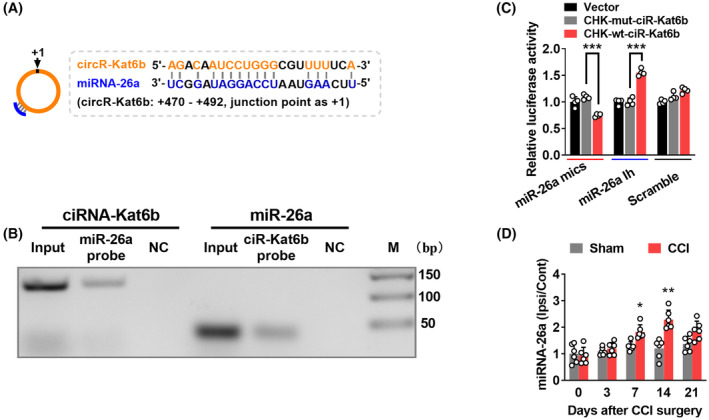
CiRNA‐Kat6b acts as competing endogenous RNAs to sponge miRNA‐26a. (A) Schematic presentation of miRNA‐26a binding to ciRNA‐Kat6b. (B) Identification of miRNA‐26a binding to ciRNA‐Kat6b: RT‐PCR detection for ciRNA‐Kat6b using the immunoprecipitated complex with miRNA‐26a (miR‐26a probe) or its mutated probe (NC) (left); or for miRNA‐26a using the immunoprecipitated complex with wild ciRNA‐Kat6b (ciR‐Kat6b probe) or its mutated probe (NC) (right). (C) The validation of ciRNA‐Kat6b negatively regulating miRNA‐26a by luciferase reporter assay in vitro. A fragment of ciRNA‐Kat6b containing the bound region by miRNA‐26a was inserted into pGL6 reporter vectors (CHK‐wt‐ciR‐Kat6b). A mutation was generated via altering the sequence bound by miRNA‐26a as indicated (CHK‐mut‐ciR‐Kat6b). The wild and mutation reporters were, respectively, co‐transfected into the HEK293T with miRNA‐26a mimics (miR‐26a mics) or inhibitor (miR‐26a Ih) or the scrambled. n = 4 per group. ***p < 0.001 versus CHK‐mut‐ciR‐Kat6b. Two‐way ANOVA (effect vs plasmid treated interaction) followed by post hoc Tukey test. (D) Levels of miRNA‐26a in the ipsilateral dorsal spinal cord and contralateral dorsal spinal cord after chronic constriction injury (CCI) or Sham surgery of unilateral sciatic nerve. n = 6 mice/time point/group. *p < 0.05, **p < 0.01 versus the corresponding Sham group by two‐way ANOVA followed by post hoc Tukey test.
