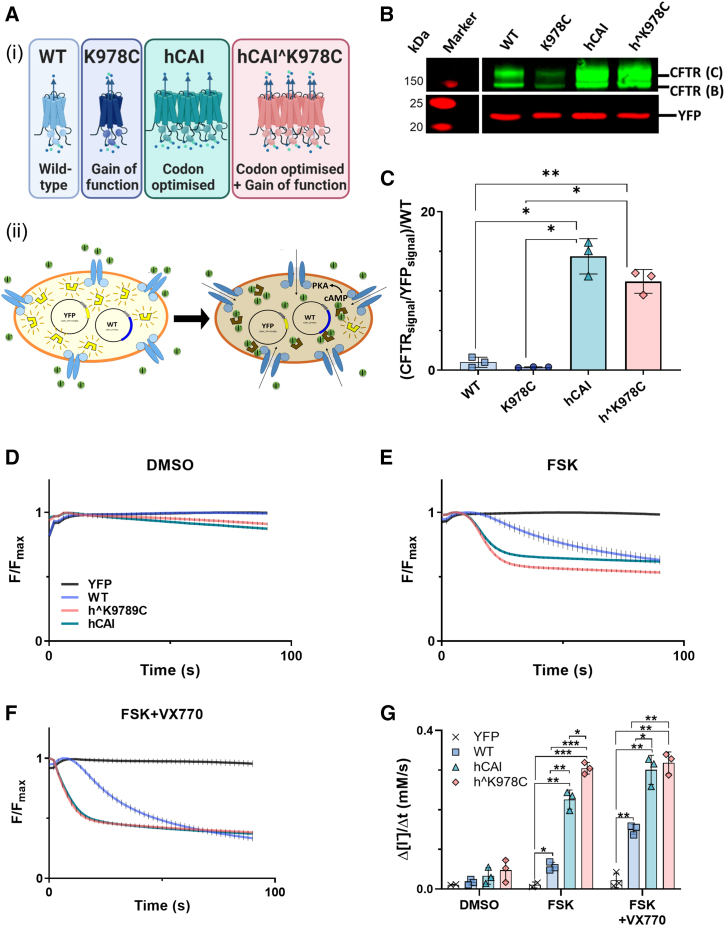
Figure 1.
Impact of CFTR cDNAs on protein production and function in transfected HEK293T cells
(A) Illustrations representing each CFTR variant (WT [blue], K978C mutation [purple], high codon adaptation index [hCAI] CFTR [cyan], and hCAI carrying the K978C mutation [hˆK978C, pink]) (i) and schematic of the halide-sensitive (HS) YFP quenching assay (HEK293T cells co-transfected with HS-YFP and CFTR variants) (ii). CFTR activators induce facilitated anion movement through CFTR channels, leading to HS-YFP fluorescence quenching. (B) Western blot of protein extracts from HEK293T cells co-transfected with HS-YFP and CFTR cDNAs. CFTR bands C and B (green) and HS-YFP (red) are indicated. Molecular weight markers (red) are displayed on the left side of the blot. (C) Quantification of CFTR protein abundance, expressed as the mean of (CFTR (bands C + B)/HS-YFP) normalized to the WT for standardization between blots. Data are represented as mean ± SD. Differences were compared by one-way ANOVA followed by Tukey’s post hoc tests. ∗p < 0.05, ∗∗p < 0.01, n = 3. (D–F) HS YFP fluorescence quenching over time in HEK293 cells transfected with HS YFP alone or co-transfected with CFTR cDNAs under different conditions: DMSO (vehicle) (D), forskolin (FSK) (E), and FSK plus the CFTR potentiator VX770 (FSK+VX770) (F), all added at time point 0. (G) The maximal rate of I− influx (Δ[I−]/Δt) is summarized for CFTR cDNA and conditions. Data are represented as mean ± SD along with individual data points. CFTR function was compared by two-way ANOVA followed by Tukey’s post hoc tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n = 3.