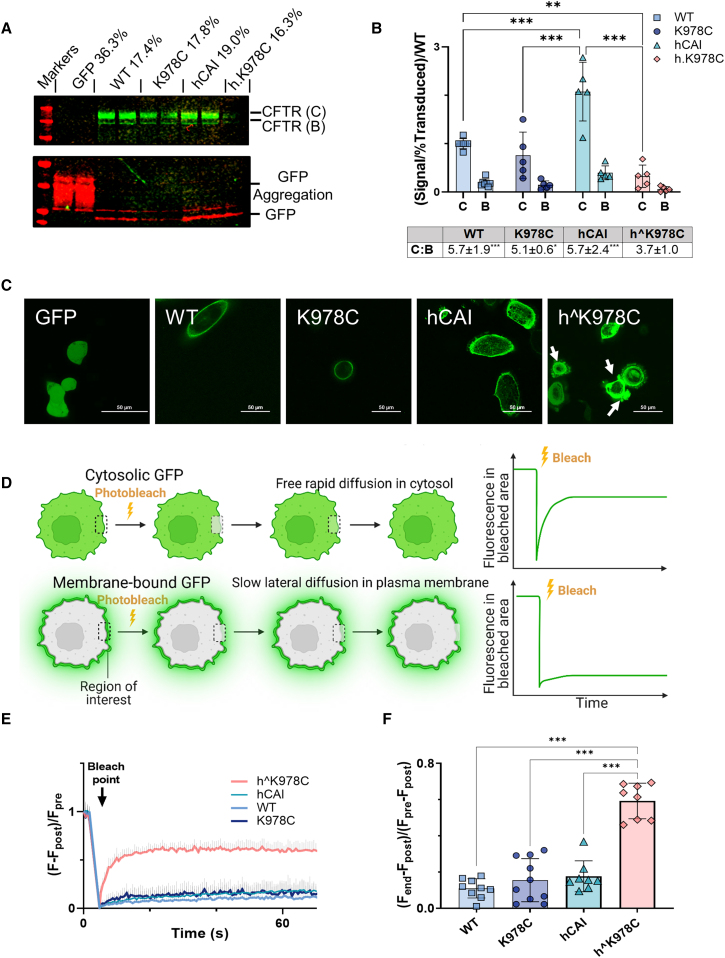
Figure 2.
CFTR protein expression and localization in CFBE cells
(A) Representative western blot of soluble protein fraction from differentiated CFBE cell cultures transduced with CFTR cDNAs or GFP alone (as labeled). Blots are probed with anti-CFTR (CFTR 596) and anti-GFP. CFTR bands C and B (green), GFP (red), and molecular weight markers (red) are displayed. (B) Summary of the relative fluorescence units (RFUs) for CFTR bands C and B in relation to the percent final transduction, normalized to the WT for each CFTR cDNA. Data are shown as mean ± SD with individual data points (n = 4–7 from 3 donors). Treatments were compared by two-way ANOVA with Tukey’s post hoc analyses. ∗∗p < 0.01, ∗∗∗p < 0.001. RFU ratios of bands B:C are displayed below the graph. (C) Representative confocal live images (×60, excitation 488 nm/emission 507 nm) of CFBE BMI-1 cells transduced with GFP linked to CFTR cDNAs or GFP alone. Cell blebbing is indicated with white arrows, with a 50-μm scale bar displayed. (D) Schematic detailing the FRAP protocol, indicating the stages of pre-bleach, post-bleach, and end, within the region of interest (ROI), represented by the white square. (E) Kinetics plot showcasing fluorescence (F) change over time in an ROI on the cell periphery. The bleach point is marked with an arrow. (F) Summary of the mobile fraction (Mf) ratio for different GFP-linked CFTRs. Data are shown as mean ± SD with individual data points. Treatments were compared by one-way ANOVA with Tukey’s post hoc analyses. ∗∗p < 0.01, ∗∗∗p < 0.001. (n = 8–13 from 1 CFBE BMI-1 cell donor: F508del/F508del).