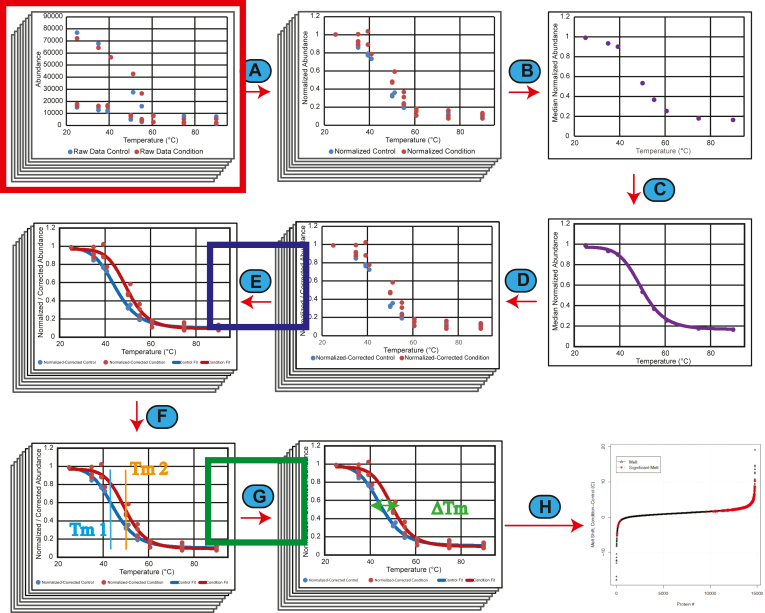
Fig. 1.
Graphical overview of the computational steps in InflectSSP.A, abundance data from each protein normalized by dividing each abundance value at each temperature by the abundance at the lowest temperature. B, median of all abundance values across proteome at each temperature and for each condition and control across all replicates. C, curve fitting of proteome data sets. D, correction of individual normalized abundance values for each protein based on difference between actual and predicted in previous step. E, curve fitting for each protein melt curve based on corrected normalized abundance values. R2 for curve fits are also calculated. F, calculation of melt temperature for each condition and control across all proteins. G, calculation of melt shift along with melt shift p-value. H, result reporting that describes the melt shifts calculated in the experiment with outputs in various formats (plots and tables). Potential locations in the data analysis workflow where quality of the data analysis can be evaluated. The red box indicates the number of peptide spectrum matches (PSMs) and number of unique peptides (UPs) that are reported with the protein/abundance data from the MS experiment and proteomic search. The blue box represents where the coefficient of determination (R2) is calculated based on the fit of the melt curves. The green box represents where the melt shift p-values are calculated in the workflow with the melt shifts.