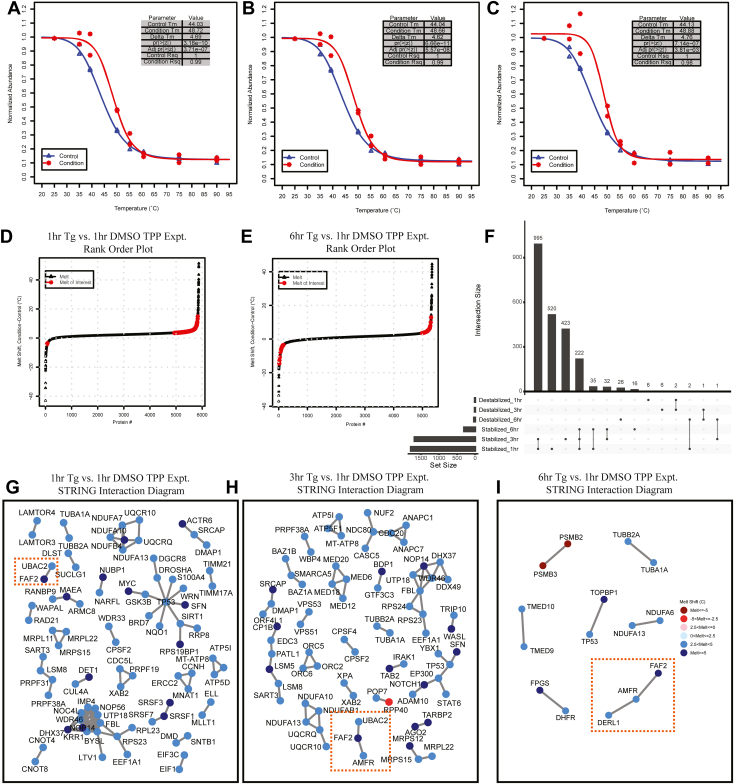
Fig. 7.
InflectSSP output from analysis of TPP time course experiments where HEK293A cells were treated with thapsigargin for 1, 3, or 6 h or DMSO for 1 h.A–C, melt curves for SERCA2A at (G) 1 h, (H) 3 h, and (I) 6 h relative to DMSO at 1 h. D and E, rank order plots for all protein melt shifts in the time course experiments where proteins with melt shift p-value <0.05, melt shifts >3.5 °C or <−3.5 °C are highlighted in red. Plots are of melt shifts calculated between (D) DMSO at 1 h versus thapsigargin at 1 h, (E) DMSO at 1 h versus thapsigargin at 6 h. F, upset plot describing overlap of stabilized and destabilized proteins in the three time points in the thapsigargin time course experiment. G–I, STRING interaction diagrams for proteins with reported interactions, melt shifts >3.5 °C or <−3.5 °C, and STRING confidence score = 0.99. Diagrams are for (G) DMSO at 1 h versus thapsigargin at 1 h, (H) DMSO at 1 h versus thapsigargin at 3 h, (I) DMSO at 1 h versus thapsigargin at 6 h. Orange boxes in all three interaction diagrams highlight ERAD protein interactions that are present in all three data sets. ERAD, ER-associated degradation process; TPP, thermal proteome profiling.