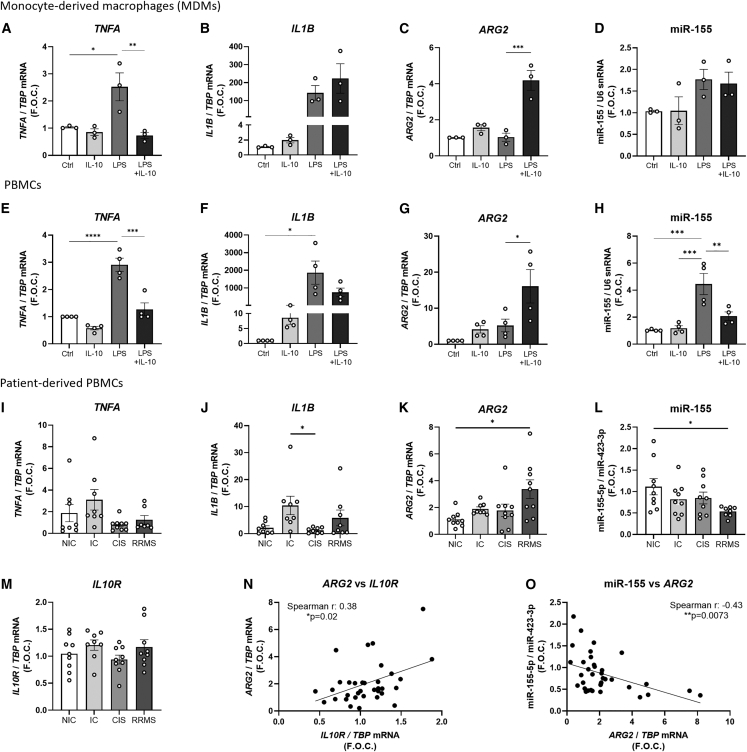
Figure 1.
Gene expression in stimulated human MDMs and PBMCs and in unstimulated patient-derived PBMCs
Expression levels of (A) TNFA, (B) IL1B, (C) ARG2, and (D) miR-155 in stimulated MDMs (n = 3) and of (E) TNFA, (F) IL1B, (G) ARG2, and (H) miR-155 in PBMCs isolated from buffy coat bags donated by healthy donors (n = 4) are shown. TBP was used as the endogenous control while U6 snRNA was used as the control for microRNA analysis and graphed as fold over control (F.O.C.). Expression levels of (I) TNFA, (J) IL1B, (K) ARG2, (L) miR-155, and (M) IL10R in four different participant groups, i.e., non-inflammatory controls (NIC) (n = 9), inflammatory controls (IC) (n = 9), clinically isolated syndrome (CIS) (n = 9), and in patients with relapsing remitting multiple sclerosis (RRMS) (n = 9) where samples were taken during the remission phase, are shown. TBP was used as the endogenous control while miR-423-3p was used as the endogenous microRNA control. Results were graphed as F.O.C. A Spearman’s correlation analysis was performed to analyze the correlation between the expression of ARG2 and (N) IL10R and (O) miR-155 in all PBMC samples. Graphs (A–H) were analyzed using a one-way ANOVA and Tukey’s multiple comparisons test and graphs (I–N) were analyzed using a Kruskal-Wallis test and Dunn’s multiple comparisons test. Correlation analysis was performed using a nonparametric Spearman correlation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars are representative of the standard error of the mean (SEM).