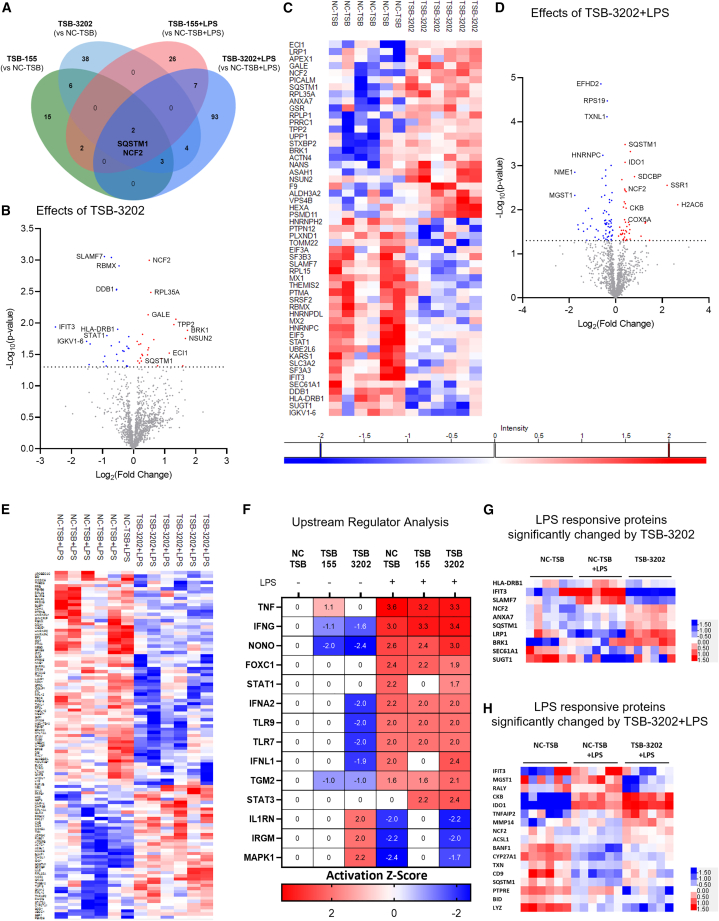
Figure 4.
Mass spectrometry-based proteomic analysis of the effects of Arg2 TSBs in human MDMs
(A) A Venn diagram was used to illustrate the number and overlap of significantly differentially expressed proteins in MDMs transfected with TSB-NC, TSB-155, and TSB-3202, in the presence or absence of LPS stimulation (n = 3 independent experiments, performed in duplicate). Group comparisons made were included on the Venn diagram. (B) Volcano plot showing the effects of TSB-3202 compared with NC-TSB based on a Log2 (fold change) and –Log10 (p value). Proteins with a significant p value of <0.05 (–Log10 p > 1.3) are highlighted in red (increased fold change) and blue (decreased fold change). (C) Heatmap representing the effects of TSB-3202 compared with NC-TSB, where LFQ intensities of significantly changed proteins are represented as Z scores. (D) Volcano plot showing the effects of TSB-3202 + LPS compared with NC-TSB + LPS based on a Log2 (fold change) and –Log10 (p value). Proteins with a significant p value of <0.05 (-Log10 p value of >1.3) were highlighted in red and blue. (E) Heatmap representing the effects of TSB-3202 + LPS compared with NC-TSB + LPS (n = 3 independent experiments, performed in duplicate). (F) All treatment conditions were compared with NC-TSB and the lists of significantly changed proteins were analyzed by Ingenuity Pathway Analysis software (QIAGEN) to determine the upstream regulators and their activation Z scores (p < 0.01). Upstream regulators with an activation Z score ≥1.6 or ≤ −1.6 modulated by NC-TSB + LPS were graphed alongside STAT-3 to highlight the upstream regulators of LPS. The corresponding activation Z scores of TSB-3202 and TSB-155 with and without LPS were included, the p value of overlap was p < 0.01. Comparison of the NC-TSB with NC-TSB + LPS was used to generate a list of LPS-responsive proteins, which was intersected with the proteins that were significantly changed by (G) TSB-3202 treatment (vs. TSB-NC) and (H) TSB-3202 + LPS treatment (vs. NC-TSB + LPS). Z scores were used to generate heatmaps. Error bars are representative of the standard error of the mean (SEM). Statistical analysis was performed on proteomic data using a Student’s t test to identify the significantly changed proteins.