Figure 5. FAM120A bridges SRSF1 to SREBP1 to induce lipogenic enzyme expression.
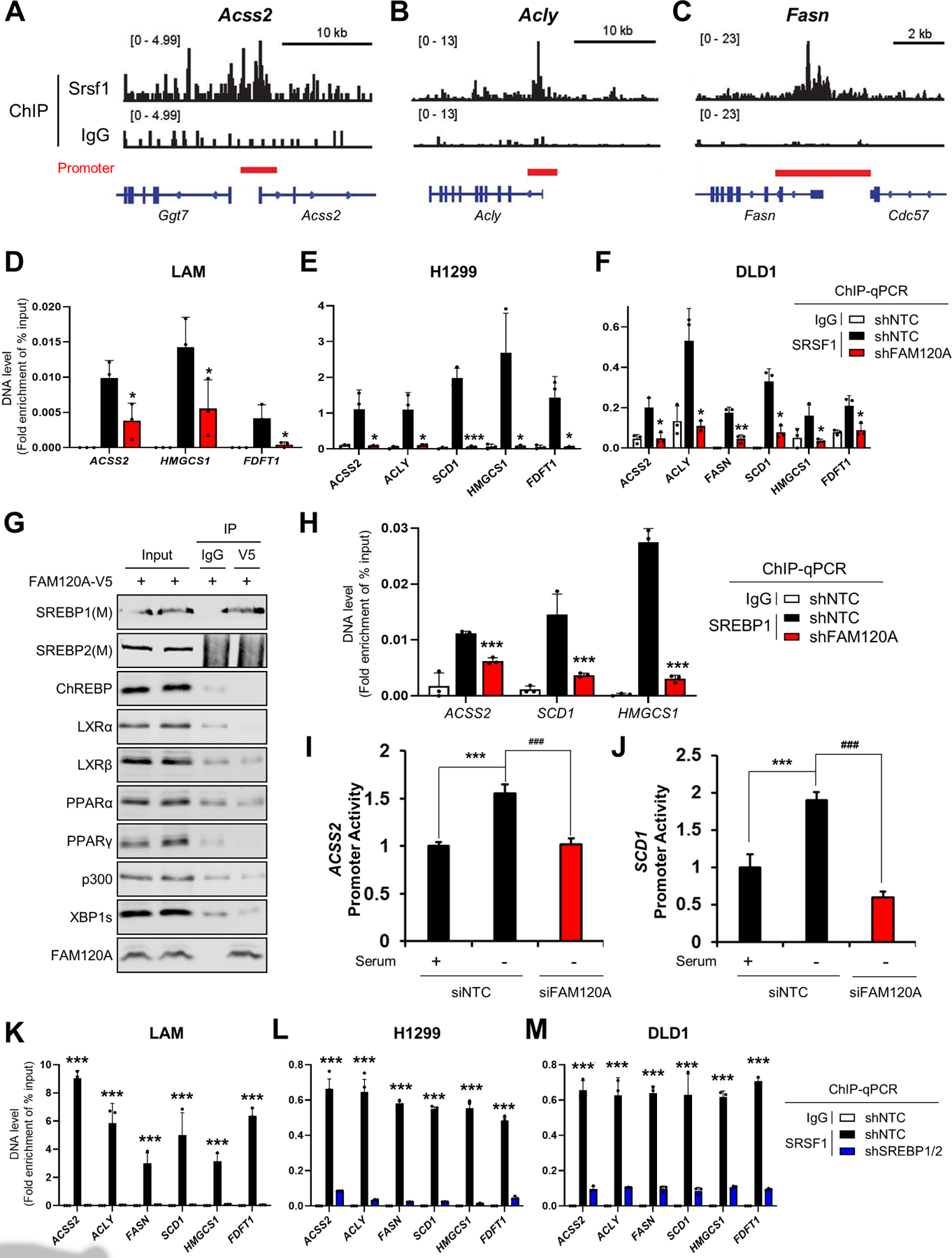
(A-C) Analysis of Srsf1 chromatin immunoprecipitation (ChIP)-seq results (GSE45517)46 in MEF cells near the transcription start sites of Acss2 (A), Acly (B), and Fasn (C).
(D-F) ChIP-qPCR analysis of SRSF1 in target gene promoters. Immunoprecipitation was performed using IgG or anti-SRSF1 antibodies in LAM (D), H1299 (E), and DLD1 (F) cells stably expressing shRNAs targeting FAM120A or control. Cells were serum-starved overnight. p-value was calculated between shNTC and shFAM120A. N = 3.
(G) Co-IP analysis of transcription factors in HEK293E cells expressing FAM120A-V5. 1% of total cell lysate was loaded as an input.
(H) ChIP-qPCR analysis of SREBP1 on target gene promoters. Immunoprecipitation was performed using IgG or anti-SREBP1 antibodies in LAM cells stably expressing shRNAs targeting FAM120A or control. Cells were serum-starved overnight. p-value was calculated between shNTC and shFAM120A. N = 3.
(I, J) Promoter activity analysis of ACSS2 (H) and SCD1 (I) in LAM cells transfected with promoter constructs and siRNAs targeting FAM120A or control. N = 3.
(K-M) ChIP-qPCR analysis of SRSF1 in target gene promoters. Immunoprecipitation was performed using IgG or anti-SRSF1 antibodies in LAM (K), H1299 (L), and DLD1 (M) cells stably expressing shRNAs targeting SREBP1/2 or control. Cells were serum-starved overnight. p-value was calculated between shNTC and shSREBP1/2. N = 3.
Data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, and ###p < 0.001.
See also Supplemental Figure S4.
