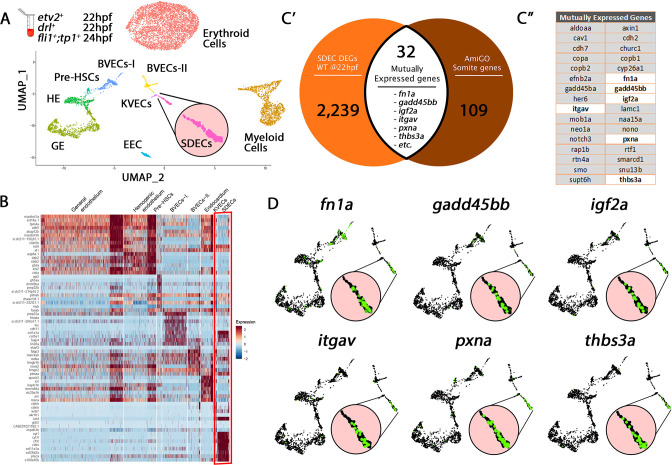
Figure 1. Cell-type-specific endothelial cell markers highlight cellular diversity within the vasculature.
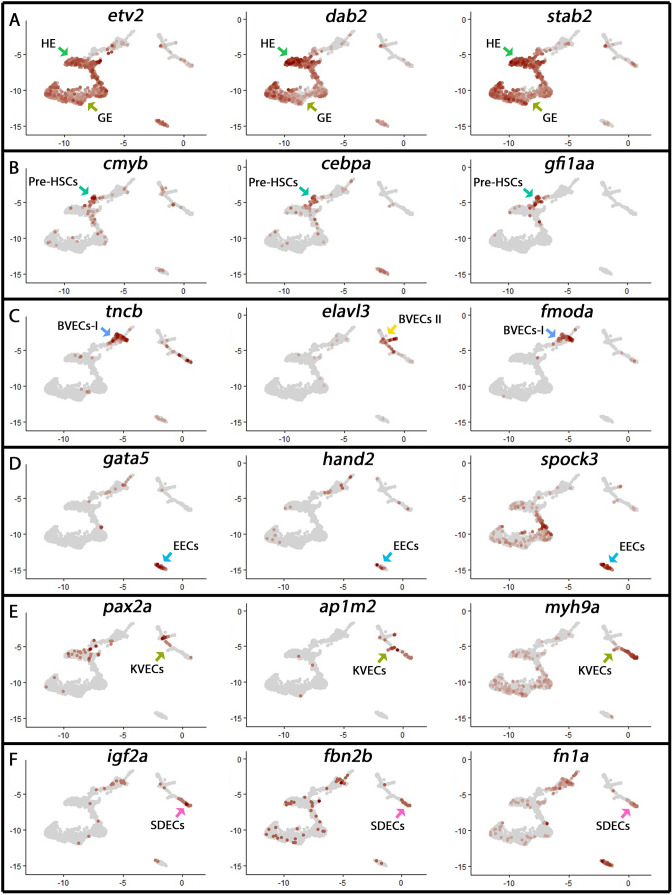
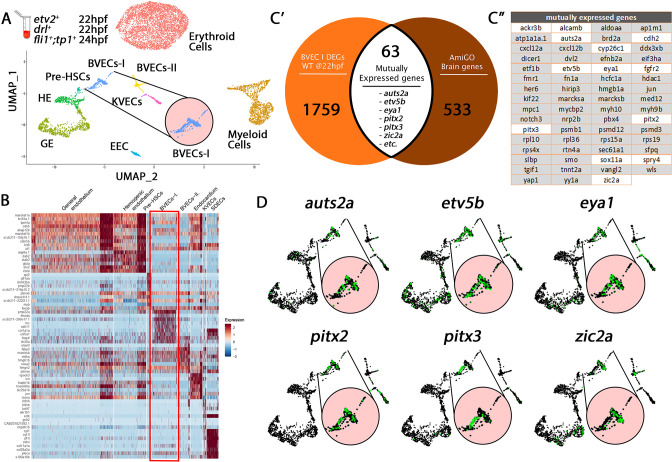
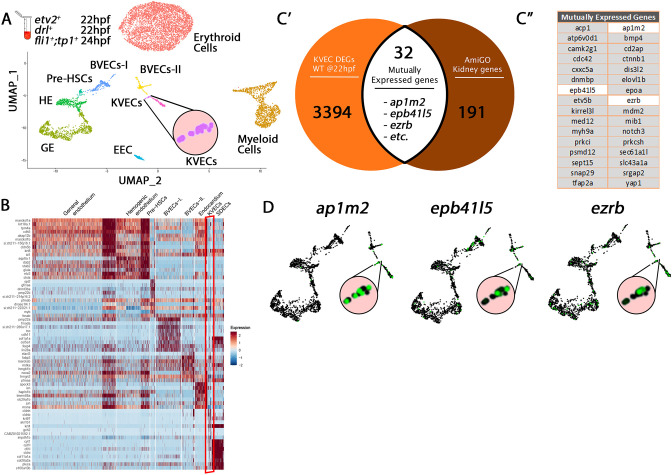
(A) Uniform manifold approximation projection (UMAP) plots of scRNA-seq data of total endothelial lineage cells collected from TgBAC(etv2:Kaede)ci6, Tg(fli1:DsRed)um13; Tg(tp1:GFP)um14, and Tg(drl:H2B-dendra) embryos at 22–24 hpf. Clusters were named according to their gene expression: Erythroid, Lymphoid, General Endothelium (GE), Hemogenic Endothelium (HE), Pre-HSCs, Brain Vascular Endothelial Cells (BVECs-I and BVECs-II), Kidney Vascular Endothelial cells (KVECs), Endocardial Endothelial Cells (EECs), and somite-derived endothelial cells (SDECs). Color-coded marker gene expression levels are shown on corresponding clusters. A pink circle highlights the SDEC cluster. (B) Expression heatmap of 22–24 hpf single-cell transcriptome shows the top predicted differentially expressed marker genes across the different clusters. A red box highlights the SDEC cluster. (C’,C”) A list of somite-annotated genes was curated from the AmiGo annotation database and compared with the SDEC transcriptome. 32 genes were commonly expressed. Interestingly, several of these 32 genes were enriched within the SDEC cluster (C”; white boxed genes, D; enlarged circles).