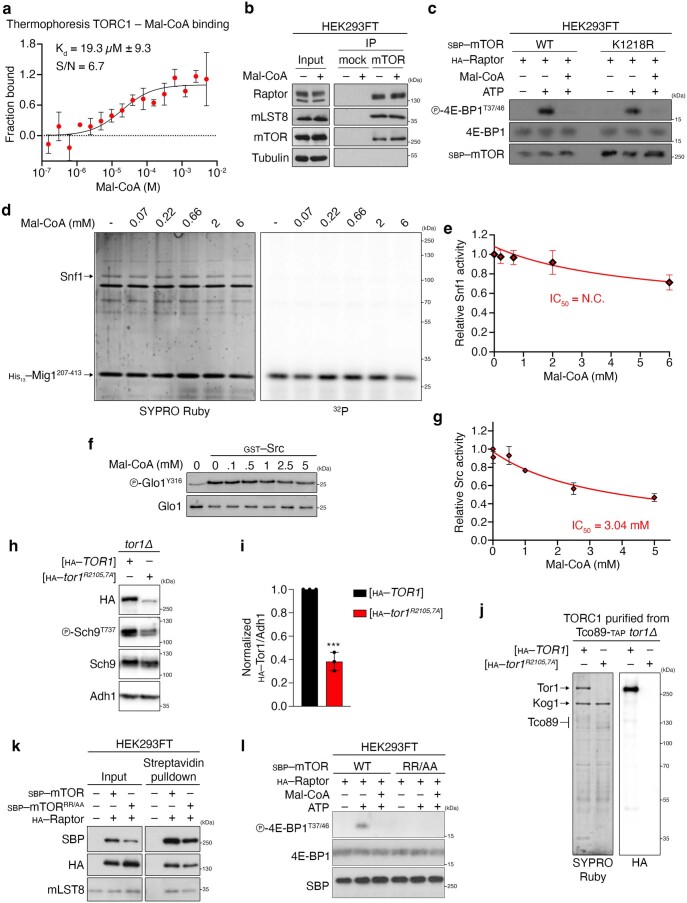
Extended Data Fig. 10. Mal-CoA binds and specifically inhibits mTORC1 without affecting complex composition and independently of mTOR malonylation, while mutant Tor1/mTOR in the Mal-CoA stabilization residues show decreased stability and/or activity.
a, Microscale thermophoresis experiment using purified TORC1 (containing GFP–Tor1) from yeast cells, confirming binding of Mal-CoA to TORC1; n = 3 independent experiments. b, Mal-CoA does not affect mTORC1 complex stability. Endogenous mTOR was immunoprecipitated in the presence or absence of 1 mM Mal-CoA (added directly in the lysates 5 min previous to addition of the antibody). Co-immunoprecipitation of Raptor and mLST8 assayed by immunoblotting; n = 2 independent experiments. c, Mal-CoA inhibits mTORC1 independently from mTOR malonylation. IVKs as in Fig. 8c using SBP-tagged WT or K1218R mutant mTOR and HA–Raptor in the presence or absence of 5 mM Mal-CoA. Reactions omitting ATP were used as negative controls; n = 2 independent experiments. d,e, Mal-CoA does not inhibit Snf1 in vitro. d, IVK assays as in Fig. 8a, but using purified Snf1 and His-tagged Mig1 as substrate, with the indicated concentrations of Mal-CoA. e, Quantification of Snf1 activity; n = 3 independent experiments. f,g, Src IVK assay as in Fig. 8c using Glo1 as a substrate, with the indicated amounts of Mal-CoA. g, Quantification of Src activity; n = 3 independent experiments. h,i, Stability of HA-tagged Tor1R2105/2107A mutant in vivo. i, Quantification of Tor1 protein levels normalized to Adh1; n = 3 independent experiments. j, In vitro stability of HA-tagged Tor1R2105/2107A mutant purified from yeast cells via TAP-mediated pulldown of Tco89; n = 2 independent experiments. k, The human SBP-tagged mTOR R2168A/R2170A mutant (mTORRR/AA) is relatively stable and binds other mTORC1 components similarly to WT mTOR. Streptavidin pulldown of WT or mTORRR/AA detecting binding to HA–Raptor and endogenous mLST8; n = 2 independent experiments. l, The mTORRR/AA mutant lacks catalytic kinase activity in vitro. IVKs as in Fig. 8c using SBP-tagged WT or mTORRR/AA and HA–Raptor in the presence or absence of 5 mM Mal-CoA. Reactions omitting ATP were used as negative controls; n = 2 independent experiments. Data are the mean ± s.d. (a) or mean ± s.e.m. (all other graphs). ***P < 0.0005. Source numerical data and unprocessed blots are provided.