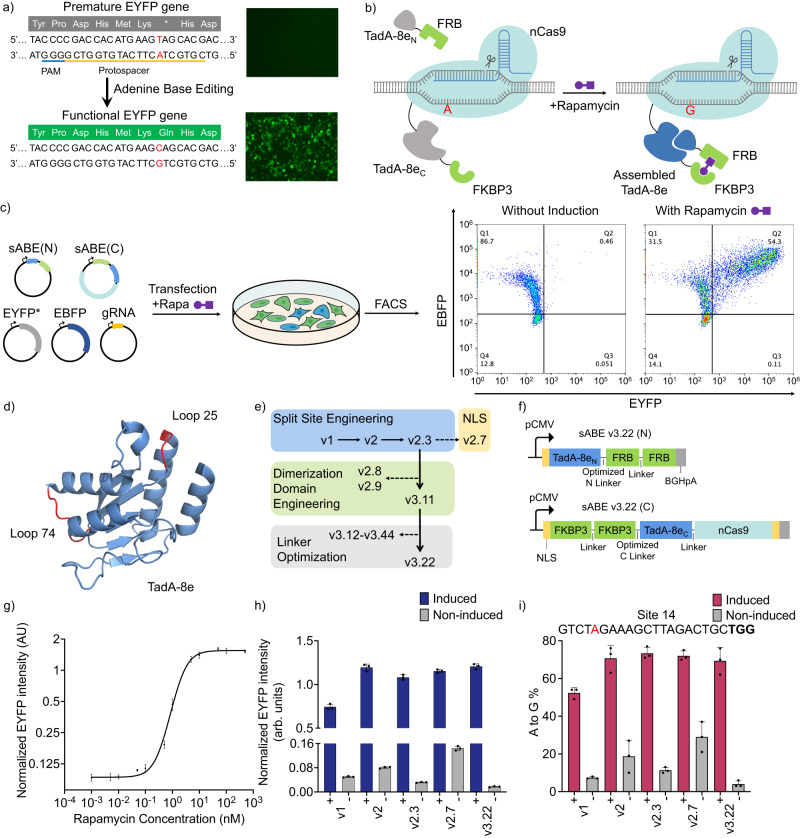
Fig. 1. Chemically inducible split ABE (sABE) with tightly regulated deaminase activity.
a Schematic of the EYFP fluorescence. An A-to-G conversion on the highlighted adenine on the antisense strand of the dysfunctional EYFP* gene can restore the expression of functional EYFP protein. b Schematic of the sABE. In the absence of rapamycin (Left), two parts of the TadA-8e: TadA-8eN and TadA-8ec, remain inactive. In the presence of rapamycin (Right), the rapamycin-FKBP3 complex binds to FRB, bringing the TadA-8eN and TadA-8ec to spatial proximity to form an active ABE unit. nCas9: Streptococcus pyogenes Cas9 (D10A) nickase; FKBP: FK506 binding proteins; FRB: FKBP-rapamycin binding domains. c EYFP* reporter assay in HEK293T cells. Cells are co-transfected with five plasmids, using EBFP as a transfection control. Representative FACS data show the EYFP* activation by sABE in the presence of rapamycin. d Crystal structure of the TadA-8e deaminase domain of ABE8e (PDB: 6VPC38). Highlighted loop-25 and loop-74 regions indicate where the TadA-8e is split into two parts for the sABE v1 and v2, respectively. e Engineering steps to increase the rapamycin-induced deaminase activity and to decrease the non-induced background. f Diagram of the sABE v3.22 constructs. pCMV: cytomegalovirus promoter. g Dosage-response curve of the reporter assay to sABE v3.22. Normalized EYFP intensity is the mean EYFP intensity divided by the mean EBFP intensity. h EYFP* reporter responses to five versions of sABEs. Blue: with 100 nM rapamycin induction; gray: non-induced. i A-to-G editing efficiencies of the highlighted adenine by five versions of sABEs at Site 14. Red: with 100 nM rapamycin induction; gray: non-induced. Editing efficiencies in (i) are evaluated by Sanger sequencing. Dots represent data from three independent biological replicates, and bars represent their mean with s.d.