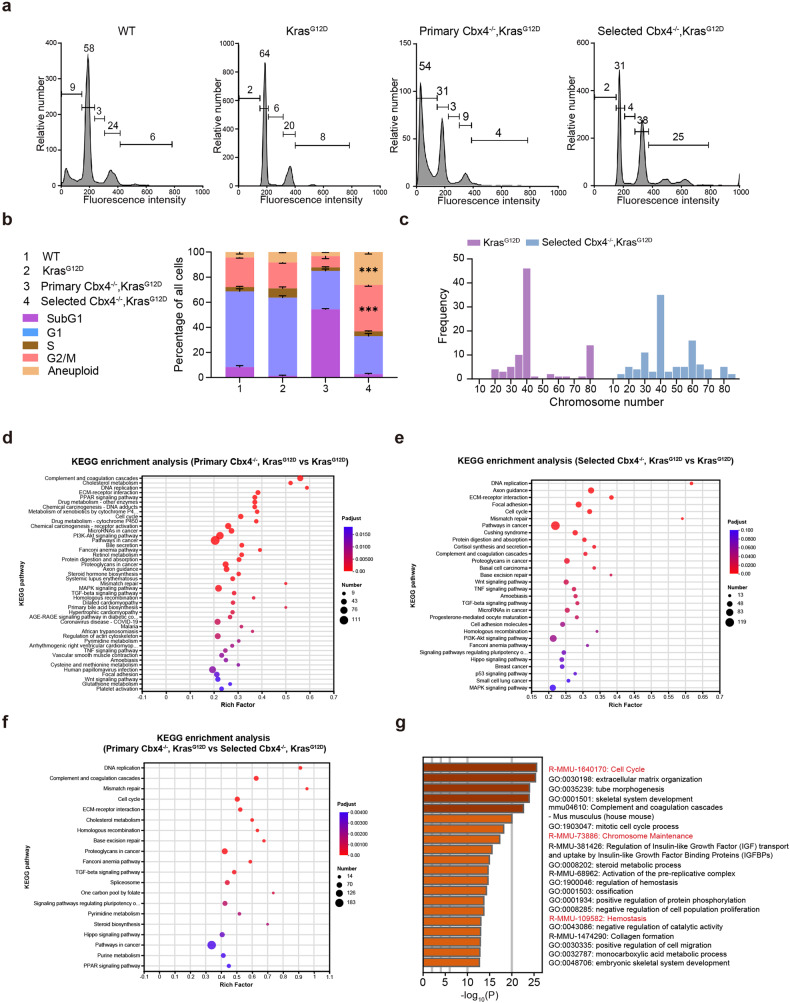
Fig. 4.
Genomic instability emerges in selected Cbx4−/−, KrasG12D MEFs compared with KrasG12D and primary Cbx4−/−, KrasG12D MEFs. a FACS Analysis of Wild-Type, KrasG12D, primary Cbx4−/−, KrasG12D and selected Cbx4−/−, KrasG12D MEFs of cell proportion at each stage. b Quantitative analysis of Wild-Type, KrasG12D, primary Cbx4−/−, KrasG12D and selected Cbx4−/−, KrasG12D MEFs of cell proportion at each stage. c Karyotype analysis of KrasG12D and selected Cbx4−/−, KrasG12D MEFs. d KEGG enrichment analysis of primary Cbx4−/−, KrasG12D MEFs compared with KrasG12D MEFs. Several cell pathways are found altered including Wnt signaling pathway. e KEGG enrichment analysis of selected Cbx4−/−, KrasG12D MEFs compared with KrasG12D MEFs. Several pathways are found altered such as Hippo, PI3K and Wnt. f KEGG enrichment analysis of primary Cbx4−/−, KrasG12D MEFs compared with selected Cbx4−/−, KrasG12D MEFs. Several cell function modules are found altered like DNA replication, pathways in cancer and Hippo signaling pathway. g GO enrichment analysis between primary Cbx4−/−, KrasG12D and KrasG12D MEFs. Data are shown as means ± SEM. ***p < 0.001