-
A
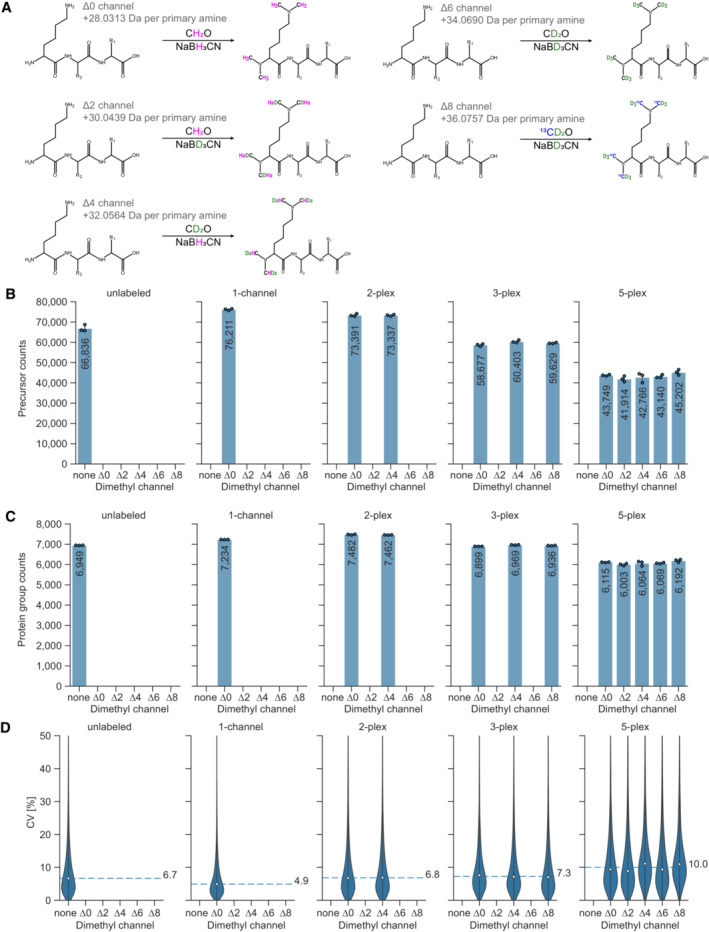
Stable isotope dimethyl labeling scheme for a five‐plex mDIA experimental setup with Lys‐N digested peptides. Depending on the combination of stable isotope labeling reagents, mass tags of 28.0313 Da, 30.0439 Da, 32.0564 Da, 34.0690 Da, or 36.0757 Da are added to each primary amine group on a peptide. Since Lys‐N hydrolyses the N‐terminal side of lysine residues, two labels are clustered on the N‐termini of peptides (N‐terminus and N‐ε‐Lys).
-
B, C
Number of quantified HeLa peptide precursors (B) and protein groups (C) for unlabeled (none), one‐channel (Δ0), two‐plex (Δ0 and Δ4), three‐plex (Δ0, Δ4, and Δ8), and five‐plex (Δ0, Δ2, Δ4, Δ6, and Δ8) labeled samples. 100 ng of peptides were injected per channel (technical replicates, n = 3).
-
D
Coefficients of variation (CV, %) of all protein groups identified per condition in the dimethyl five‐plex setup, using Lys‐N as protease. Protein group intensities are calculated using MaxLFQ‐based protein quantification from ‘Precursor.Normalised’ quantities. Median CVs for three technical replicates (n = 3) are shown as dashed lines.