Figure 4. Streamlined and automated ultra‐high sensitivity mDIA workflow.
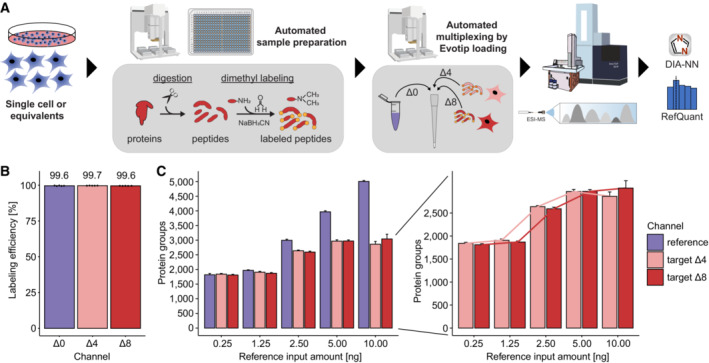
- Single HeLa cells or single‐cell equivalents are processed in a standard 384‐well plate on a Bravo robot (Agilent) by lysis in TEAB and ACN, tryptic digestion, dimethyl labeling, and multiplexing by loading the different mDIA channels onto the same Evotip. LC–MS analysis is done by an Evosep One with low‐flow chromatography coupled to a timsTOF SCP instrument. The data is analyzed by DIA‐NN, followed by our algorithm RefQuant (see main text).
- 1 ng tryptic peptides from HeLa cells were labeled with dimethyl mass tags Δ0, Δ4, and Δ8 and acquired individually in DDA mode to determine labeling efficiency. Efficiency is calculated based on intensity ratios of labeled peptides relative to all detected peptides. The labeling efficiencies were consistently higher than 99.5% for all channels in quintuplicate measurements (technical replicates, n = 5).
- Effect of varying the protein input in the reference channel for protein identification across all channels. Increasing the total protein amount in the reference channel linearly increases protein identifications in the reference channel (left), but importantly protein identifications reach a plateau in the target channels (Δ4 and Δ8) with single‐cell equivalents upon 5–10 ng in the reference channel (scReference dataset). Connected lines between increasing reference input amounts show a sigmoidal relation (right). Error bars represent the standard deviation of quintuplicate measurements (technical replicates, n = 5).
