Abstract
Background
Seasonal variations in environmental exposures at birth or during gestation are associated with numerous adult traits and health outcomes later in life. Whether DNA methylation (DNAm) plays a role in the molecular mechanisms underlying the associations between birth season and lifelong phenotypes remains unclear.
Methods
We carried out epigenome-wide meta-analyses within the Pregnancy And Childhood Epigenetic Consortium to identify associations of DNAm with birth season, both at differentially methylated probes (DMPs) and regions (DMRs). Associations were examined at two time points: at birth (21 cohorts, N = 9358) and in children aged 1–11 years (12 cohorts, N = 3610). We conducted meta-analyses to assess the impact of latitude on birth season-specific associations at both time points.
Results
We identified associations between birth season and DNAm (False Discovery Rate-adjusted p values < 0.05) at two CpGs at birth (winter-born) and four in the childhood (summer-born) analyses when compared to children born in autumn. Furthermore, we identified twenty-six differentially methylated regions (DMR) at birth (winter-born: 8, spring-born: 15, summer-born: 3) and thirty-two in childhood (winter-born: 12, spring and summer: 10 each) meta-analyses with few overlapping DMRs between the birth seasons or the two time points. The DMRs were associated with genes of known functions in tumorigenesis, psychiatric/neurological disorders, inflammation, or immunity, amongst others. Latitude-stratified meta-analyses [higher (≥ 50°N), lower (< 50°N, northern hemisphere only)] revealed differences in associations between birth season and DNAm by birth latitude. DMR analysis implicated genes with previously reported links to schizophrenia (LAX1), skin disorders (PSORS1C, LTB4R), and airway inflammation including asthma (LTB4R), present only at birth in the higher latitudes (≥ 50°N).
Conclusions
In this large epigenome-wide meta-analysis study, we provide evidence for (i) associations between DNAm and season of birth that are unique for the seasons of the year (temporal effect) and (ii) latitude-dependent variations in the seasonal associations (spatial effect). DNAm could play a role in the molecular mechanisms underlying the effect of birth season on adult health outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-023-01542-5.
Keywords: PACE, Meta-analysis, Birth season, DNA methylation, Differentially methylated regions (DMR), Latitude
Background
Plants and animals adapt to altered seasonal cues such as temperature and light by regulating their transcriptional programmes. It has been well established in humans that many traits, including physiological (e.g. blood pressure and cholesterol), behavioural traits (e.g. conception, suicidal tendencies), and life span, display distinct seasonal patterns [1–5]. The incidence of complex diseases such as cardiovascular and autoimmune disease, and psychiatric disorders shows seasonal fluctuations [6–9]. Several studies have demonstrated an association between season of birth and development of disease both in childhood and later in life. These adverse events include asthma and allergy-related diseases [10–13], neonatal immune development [14], multiple sclerosis [15] and schizophrenia [9, 16], suggesting long-lasting effects on a broad range of characteristics. Exposures that vary seasonally such as outdoor temperature, humidity, ultraviolet (UV) light, pollen levels, Vitamin D, melatonin levels, air pollution and availability of nutrients have been hypothesised to drive the season-of-birth-linked disease phenotypes (reviewed in [17]). Uncovering the molecular mechanisms underlying the effect of season of birth on lifelong phenotypes can help elucidate the biological basis for the observed associations.
Epigenetic modifications, including alterations in DNA methylation (DNAm), play a key role in the gene regulation of a variety of cellular processes. Such modifications may also act as molecular mediators between season-linked environmental factors and health outcomes. In plants and animals, the role of DNAm in the regulation of several seasonally associated genes is well documented [18–20]. In humans, the seasonal periodicity of gene expression profiles has been demonstrated for genes involved in immunity and physiology [21]. In addition, there is evidence to suggest that environmental factors alter DNAm levels in a season-specific manner in healthy individuals [22]. It is proposed that these changes in DNAm profiles may influence health outcomes later in life. Taken together, a growing body of evidence suggests a role for differential DNAm in the exposure-induced changes in gene expression and disease [23]. However, studies on the seasonality of DNA methylation at birth in humans have not assessed variations across a wide range of geographies on a large scale.
Early life environmental exposures result in specific changes in DNAm that may persist throughout the life course. For example, maternal smoking in pregnancy results in a specific DNA methylation signature in offspring [24] that persists well into adulthood [25]. Similarly, it is plausible that DNAm changes associated with birth season, a proxy for in utero exposures, could be a mechanistic pathway underlying the effect of season of birth on later life health and disease. Lockett et al. tested the effect of season of birth on the risk of allergic outcomes in adulthood as well as the association between season of birth and differential DNAm at birth, and at age 18 [26]. They demonstrated a season-associated pattern of DNAm at some CpG sites associated with apoptotic genes as well as season-dependent risk patterns of allergic diseases such as eczema and rhinitis at 18 years of age. The study was limited in sample size (N = 175) but was validated in a small independent cohort.
Here, we report the results of a meta-analysis of epigenome-wide association studies (EWAS) from 27 independent cohorts (N = 12927 participants) from the Pregnancy And Childhood Epigenetics (PACE [27]) consortium. Blood DNAm data at birth and from children aged between 1 and 11 years were used to investigate the association between season of birth and differential blood DNA methylation at these two time points. In addition, we examined if the latitude where the children were born influenced the season-of-birth-associated DNAm profiles as exposures such as sunlight, relative humidity, pollen count, infectious agents and air pollution can vary with latitude in each season.
Methods
This meta-analysis study intended to investigate the epigenome-wide associations of season of birth and DNAm at two time points, at-birth, and childhood, on a large-scale using data from multiple cohorts across differing geographies.
Cohort participants
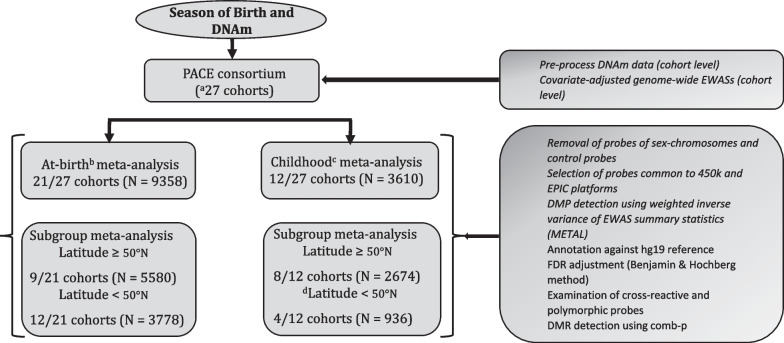
Cohorts participating in the PACE consortium with data on season of birth and DNAm at birth and/or later in childhood were invited to participate in a meta-analysis of associations between season of birth and DNA methylation. Twenty-one independent cohorts (N = 9418) contributed to the at-birth and 12 cohorts (N = 3610) to the childhood meta-analyses. Most cohorts contributed data either for at-birth or childhood analysis (21/27), while six cohorts (~ 18% of the at-birth data and ~ 45% of the childhood data) contributed to both time points. Less than 20% of the study population had data at two time points from the same child. Data for 60 individuals (EARLI, non-European) were excluded from the at-birth analysis due to insufficient numbers (as per the PACE recommendations of ≥ 15/group) of births in the spring season (n (spring) = 4, Additional file 1: Table S1B) resulting in a total of 9358 for the at-birth analysis (Fig. 1). All study participants originated from countries in the northern hemisphere (32.7–71.2°N). Additional file 1 (Table S1A-C) lists all cohorts along with the distributions of covariates used in the cohort-specific EWASs. Additional file 2 provides cohort-specific definition of covariates, DNAm data collection and pre-processing methods.
Fig. 1.
Schematic diagram for the season of birth-DNA methylation (DNAm) analyses. Numbers included in the analyses are indicated. PACE Pregnancy And Childhood Epigenetics, EWAS Epigenome Wide Association Study, DMP Differentially Methylated Probe, FDR False Positive Rate, DMR Differentially Methylated Region. EWASs and meta-analysis were carried out on at-birth and childhood samples separately. All participating cohorts were from the northern hemisphere (range 37.2–71.2°N). Latitude ≥ 50°N represents a subset of cohorts from 50 to 71.2°N (referred to as “higher latitude subgroup” in the text). Latitude < 50°N is a subset of cohorts from 32.7 to 50°N (referred to as “lower latitude subgroup” in the text. a27 independent cohorts, but a total of 33 datasets as 6 cohorts contributed to both at-birth and childhood analyses. bAt-birth blood samples (cord and heel prick). cWhole blood samples (age 1–11 years). dThis analysis was not done due to its small sample size
Exposures and outcomes
Primary analysis
The outcome was DNAm at birth and childhood. For new-born samples, all but two cohorts used cord blood, while CBC and CHS used dried blood spots collected post-delivery (Additional file 1: Table S1). The primary exposure was season of birth as a categorical variable with autumn as the reference season. Seasons were defined as winter (December–February), spring (March–May), summer (June–August) and autumn (September–November) since all participants were from the northern hemisphere.
Secondary analysis
Associations of season of birth and DNAm at-birth were also investigated in samples stratified by the cohort latitude to explore its influence on DNAm signals. Cohorts were dichotomized into a “lower latitude subgroup” (32.7–50°N with N = 3838) and “higher latitude subgroup” (≥ 50°N with N = 5580). The choice of 50° as the cut off was arbitrary, although it provided ~ 8 h of sunlight/day at the winter equinox. We focussed on comparing (i) the impact of latitudes on the pattern of association between season of birth and methylation signals at-birth and (ii) the pattern of season of birth specific associations across the two time points (at birth and in childhood) in each latitude. However, due to the small sample size contributing to childhood DNAm analysis in the lower latitude (N = 936), the DNAm results of the child cohorts in the higher latitude subgroup alone were used for comparison with the at-birth DNAm associations.
Measurement and pre-processing of DNA methylation
Details of cohort-specific methylation assays, data handling, quality control and normalisation are given in Additional file 2. Briefly, DNAm assays were carried out on bisulfite-converted genomic DNA extracted from either cord blood/heel prick samples collected at delivery or shortly after delivery and from whole blood samples from children aged 1–11 years. Methylation data for these samples were generated using illumina Infinium® HumanMethylation450 (most cohorts) or EPIC BeadChip assay (EAGeR, IoWF2, BiB of HELIX, (Additional file 1: Table S1A). All cohorts estimated methylation levels as beta values (β) using the formula where M and U are methylated and unmethylated signal intensities, respectively. Normalised beta values were used in preference to M values to afford direct comparison of our findings with similar findings in the literature where most epigenetic association studies employ beta values. While all cohorts followed a pre-specified analysis plan including recommendations for bioinformatics pipelines (Additional file 3), the individual cohorts conducted probe filtering, normalisation of methylation data and correction for batch effects using their own preferred methods. Probes on sex chromosomes and control probes were removed prior to meta-analysis. Cross-reactive probes and polymorphic probes were dealt with post-meta-analysis. Each cohort also provided information on genomic inflation in their cohort-specific EWAS models. Leave-one-out analyses were carried out to check if inflation in any one cohort unduly influenced the overall meta-analysis results.
Covariates
All cohort-specific models, at birth or childhood, included the following covariates: maternal smoking status during pregnancy, maternal socio-economic status (SES), maternal age and gestational age at delivery, new-born’s sex, and estimated cell type proportions. These covariates have been reported to be associated with either season of birth or DNA methylation making them potential confounders of the association between season of birth and DNA methylation or both [26, 28–30]. Cell type proportions were estimated by applying the Houseman method [31] to methylation data using cord blood [32] or peripheral blood reference panels [33] for at-birth and childhood analyses, respectively, as these reference-based methods were reported to be the best available methods for studies with large sample sizes [34]. In addition, the childhood models included children’s age and season of sample collection if data were available. Most cohorts used three categories for maternal smoking status during pregnancy (no smoking, quit early during pregnancy and smoked throughout pregnancy), but cohorts lacking this level of detail or with too few who smoked during pregnancy used any versus no smoking. The definition of maternal SES was cohort specific and was mainly based on maternal education, occupation and/or income. Ninety-six percent of the study population were of European ancestry despite being drawn from multiple countries. Ethnicity was, therefore, not controlled for in the models. Cohort-specific covariate data collection methods are described in Additional file 2. In general, most cohorts collected data through questionnaires/interviews and/or from medical records. Individual cohorts used their preferred batch correction methods, and they typically comprised adjustments for methylation profiling-specific variables such as plates, bead chip or other relevant technical covariates. Studies which used a sampling scheme included the sampling variable as a covariate.
Statistical methods
Cohort-specific epigenome-wide association studies (EWAS)
Each cohort performed independent EWAS analysis following a pre-specified analysis plan and an R script (Additional file 3). The primary exposure variable, season-of-birth, was consistently coded by all cohorts with autumn as the reference season. Data on one child per family were included (randomly selected) in the cohort-specific EWAS if multiple new-borns/siblings from the same family were enrolled in the cohort. Only samples with complete data for all covariates were included in EWAS analyses.
Cohort-specific EWASs modelled the association between season of birth (exposure) and DNAm (outcome) at a CpG site of the at-birth or childhood blood DNAm data using analysis of covariance via robust regression applying Huber’s weighting, implemented in the R function rlm(). Birth seasons were defined as winter (December–February), spring (March–May), summer (June–August) and autumn (September–November). Briefly, cohorts generated a regression model comparing children born in spring, summer, and winter against autumn as the reference for each of the two time points (at-birth or childhood for which they had methylation data). Models were adjusted for cohort-specific covariates as described above and in Additional file 2.
Meta-analyses
Meta-analyses were carried out using fixed effect inverse variance weighted option of METAL [35]. Meta-analyses for at-birth and childhood data were performed separately. All analyses were repeated by one other member of the team independently. Probes that were common to both EPIC and 450 k platforms alone were included in the meta-analysis [36]. A total of 470870 probes were retained after the meta-analyses of EWAS summary results. Each meta-analysis result was checked visually using quantile–quantile (QQ) plots and quantitatively by estimating lambda values, an indicator of inflation. Bias and inflation were estimated following the BACON method of van Iterson et al. [37]. Meta-analysed probes were annotated against hg19 reference genome and mapped to the nearest genes and genomic locations (CpG islands, shores, shelves) using the Illumina 450k manifest or ACME R package [38] when not available in the manifest. The annotated output from meta-analysis holds information on probes which overlap SNPs or are within 10 bases from the CpG sites (polymorphic probes). These were disregarded while interpreting the significantly differentially methylated CpGs. Associations were considered significant for CpGs that reached a statistical significance based on a false discovery rate (FDR) below 5% [39]. The FDR-corrected probes were also checked for a more stringent Bonferroni criterion of satisfying a p value threshold of 1.06 × 10−7.
Differentially methylated regions
Differentially methylated region (DMR) analyses were carried out using comb-p in R [40] on the meta-analysed single-CpG EWAS results separately for at-birth and childhood samples. CpGs within 1000 base pairs were combined to define a region [41]. The significance threshold was set to the Šidák-corrected p value of 0.05 in all DMR analyses.
Trait enrichment analysis
Trait enrichment analysis of top CpGs (raw p values for association in meta-analyses < 1.0 × 10−5) from the primary and latitude analyses was carried out using EWAS Atlas [42]. Traits were considered significant if the p values for their odds ratios were less than 0.05.
Metastable epialleles
Fisher’s exact test was carried out in R (3.5.1) to compare the proportion of metastable epialleles amongst significant loci (p value < 0.05) in each of the birth seasons of new-born infants against the proportion in array background. The comparator was 2408 metastable epialleles listed in three separate studies [43–45].
Results
Study population
Twenty-seven studies from the PACE consortium participated in the meta-analysis study to investigate possible associations between season of birth and DNAm at birth and in childhood [at-birth: 21 studies, children: 12 studies of which six cohorts contributed to both (Fig. 1, Additional file 1: Table S1A)]. There was a total of 9358 and 3610 participants for the at-birth and childhood analyses, respectively (Fig. 1). A subgroup analysis, carried out on participants living in regions of ≥ 50°N, included 5580 new-borns (9 studies) and 2674 children aged 1–11 years (8 studies) [see Additional file 1: Table S1 (A-C)].
The births were evenly distributed across the seasons (22–27.5%, Additional file 1: Table S1B-C). Approximately 50% of the babies and children included in the analyses were girls. The median maternal age at delivery was 30 years [range (min–max): 24–34.2] for the participants of the at-birth analyses and 30 years (range 26.7–32.1) for the childhood analyses. The median gestational age was 39.5 weeks (range 38.9–40.2) and 39.6 (range 36.7–40.2) for the at-birth and childhood participants, respectively. The proportion of mothers who smoked during pregnancy ranged from 0 to 15% in most cohorts in both the at-birth and childhood samples except for the mothers of IoW F2 (36.7%) and NEST (23.2% and 20% for the two ethnic groups) for the at-birth analyses.
Association between season of birth and DNA methylation at birth and in childhood
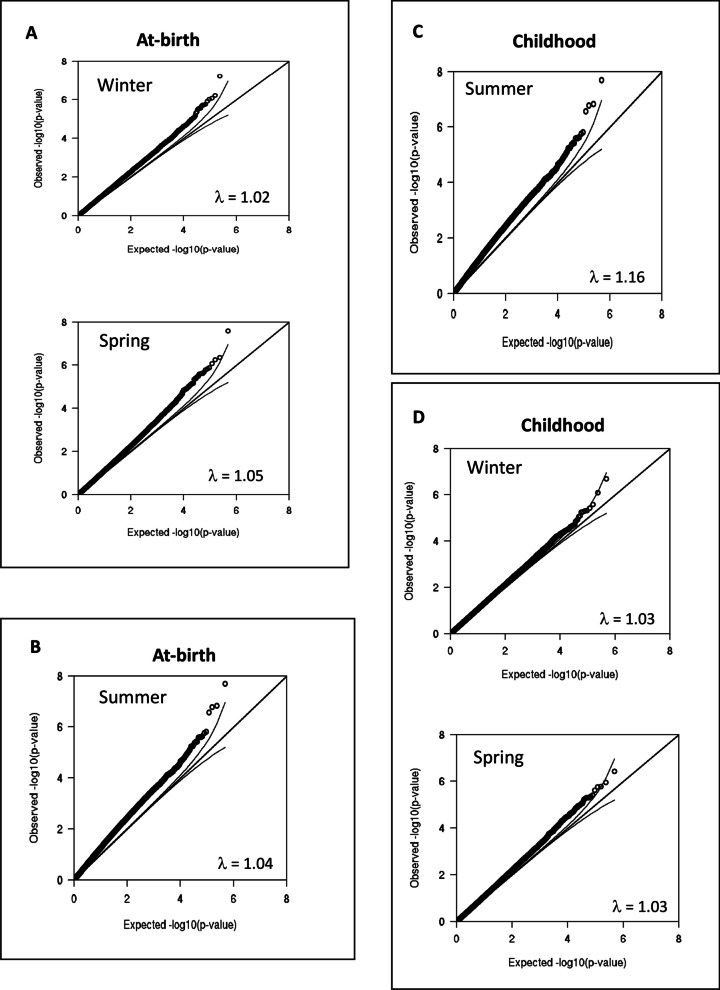
For each time point, at-birth and childhood, the individual cohorts generated an EWAS model (adjusted for covariates) comparing the three seasons (winter, spring, and summer) with autumn as a reference. The lambdas for the six models varied between 1.02 and 1.16 (Additional file 4: Table S2). Meta-analyses were then carried out on the summary results from the cohort-specific EWAS models for each of the time points (i) to identify at-birth DNAm signals associated with season of birth (winter, spring, or summer with autumn as the reference season) and (ii) to investigate whether such signals persist into childhood. Leave-one-out analyses showed that none of the individual cohorts unduly influenced the meta-analysis results.
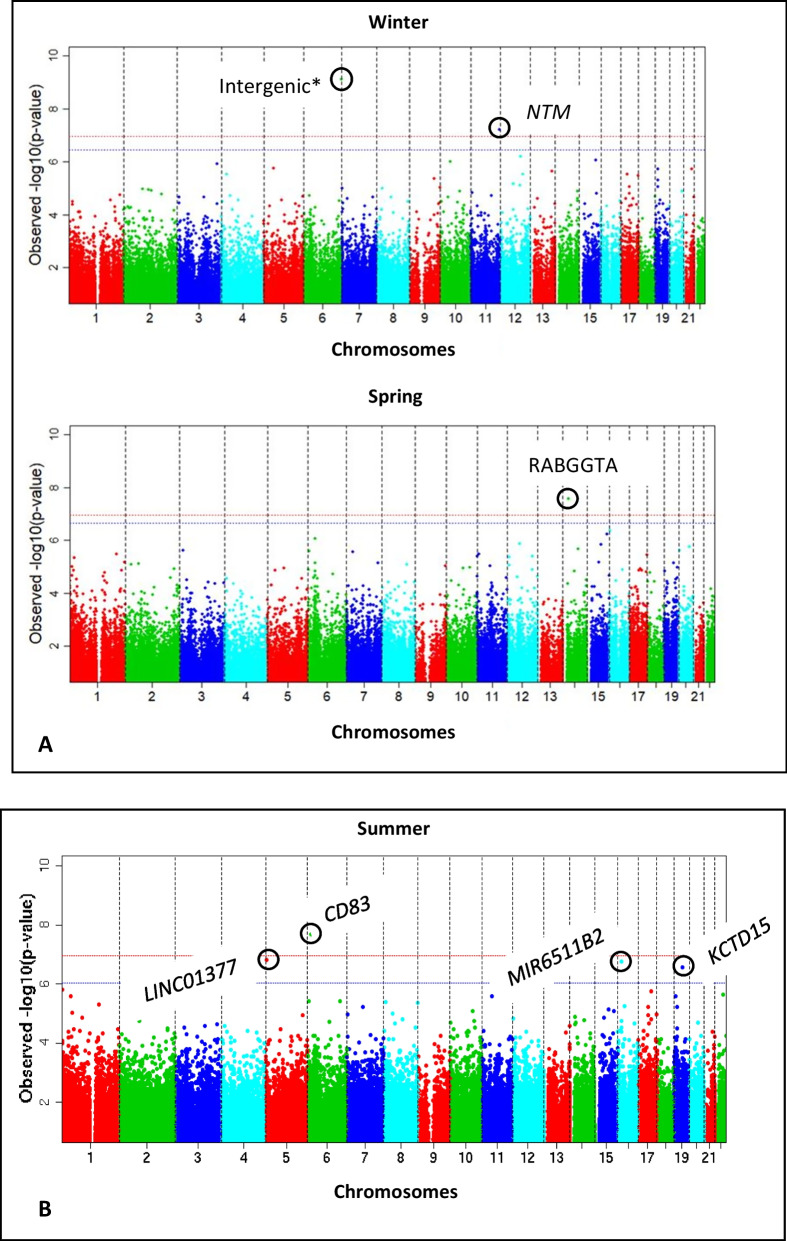
The six CpGs that passed the pre-specified 5% FDR threshold for winter-, spring- and summer-born children for the at-birth and childhood analyses are presented in Table 1 and Figs. 2 and 3. The two FDR significant CpGs identified in the new-borns, cg26416241 (winter-born, mapped to NTM) and cg18978324 (spring-born, mapped to RABGGTA), had statistical significance that passed the more stringent Bonferroni-corrected threshold of 1.06 × 10−7 (CpGs shown in bold, Table 1). The methylation levels of the at-birth CpGs were higher in the winter- and spring-born offspring) when compared to those born in the autumn. This trend was consistent across most of the cohorts (winter: 18; spring: 17 out of a total of 24 cohorts). There was no statistical evidence to suggest that the two CpGs (cg26416241, cg18978324) identified in the at-birth analysis (preliminary analysis) persisted into childhood (Additional file 4: Table S3). In the individual cohort samples of the relevant seasons, these two CpGs mostly had higher methylation levels when compared to the autumn levels in both at-birth samples (75–77% of the cohorts) and childhood samples (58% for both CpGs).
Table 1.
Differentially methylated CpGs (FDR < 0.05) in the birth seasons in at-birth and childhood analyses
| Season of birtha | CpG | Coeff | SE | p value | FDR p value | Lambdab | CHR | Position | Mapped gene | Directionc | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| At-birthd | |||||||||||
| Winter | cg26416241 | 0.0034 | 0.0006 | 6.02 × 10−8 | 0.014 | 1.02 | 11 | 131557414 | NTM | + + + − + + + + − + − + + + + − + + − + + + + − | 42.5 |
| Spring | cg18978324 | 0.0006 | 0.0001 | 2.61 × 10−8 | 0.012 | 1.05 | 14 | 24740937 | RABGGTA | − + + + + + + ? + − + + + + + + ? + − + − + + − | 27 |
| Childhood | |||||||||||
| Summer | cg19416462 | − 0.0037 | 0.0007 | 1.99 × 10−8 | 0.009 | 1.16 | 6 | 14533530 | CD83 | − − − − − − − + − − − − | 45.9 |
| cg01656588 | − 0.007 | 0.0013 | 1.47 × 10−7 | 0.026 | 5 | 3187100 | LINC01377 | − − − − − − − − − − − − | 0 | ||
| cg03263237e | − 0.0157 | 0.003 | 1.67 × 10−7 | 0.026 | 16 | 15227611 | MIR6511B2 | − − ? − − − ? − − ? + ? | 24.6 | ||
| cg15437053 | 0.0037 | 0.0007 | 2.69 × 10−7 | 0.032 | 19 | 34311123 | KCTD15 | + ? + + + + + + + + + + | 32.9 | ||
CpGs shown passed the FDR (False discovery rate) p value threshold of 5% in meta-analyses of EWAS summary results carried out with 21 and 12 cohorts for at-birth and childhood blood samples, respectively. The p values for the CpG sites that also passed the Bonferroni-corrected significance threshold of 1.06 × 10−7 are shown in bold. All cohort-specific EWAS analyses were adjusted for sex of the child, gestational age at delivery, maternal age at delivery, maternal smoking during pregnancy, maternal socio-economic status, batch, child’s age at the time of sample collection (in the childhood analyses) and estimated cell proportions
Coeff: regression coefficient (change in mean methylation compared to autumn reference); SE: standard error of the coefficient; Lambda: genomic inflation factor; CHR: chromosome; I2: a measure of heterogeneity between studies
aReference season for the EWAS analyses of individual cohorts: autumn
bEstimated using the BACON method (van Iterson et al. [37])
cHyper-methylation is indicated by “+” and hypo-methylation by “−” (minus sign)
dOne CpG (in winter-born; cg01801443; p value: 7.77 × 10−10) with a SNP within 10 base pairs of the probe is not shown in the table
eCross-reactive probe (Chen et al. [46])
Fig. 2.
Manhattan plots of the meta-analysed data showing the association of season of birth and DNA methylation at birth using neonatal blood data (A) and during childhood using whole blood data (B). Birth seasons with FDR-significant CpGs when compared to autumn as the reference season alone are shown in the figure. The blue and red lines indicate the threshold p values for false discovery rate (FDR) and the Bonferroni adjustments, respectively. The observed p values on the Y-axis are from models adjusted for covariates and cell types for the seasons indicated in the figures when compared to autumn as the reference season. Genes associated with the CpGs (circled) are indicated. All cohort-specific EWAS analyses were adjusted for gender of the child, gestational age at delivery, maternal age at delivery, maternal smoking during pregnancy, maternal socio-economic status, batch, child’s age at the time of sample collection (in the case of childhood samples and if data were available) and estimated cell proportions. *This CpG (cg01801443, location: intergenic) has a SNP within 10 base pairs and is not included in Table 1
Fig. 3.
Quantile–quantile (Q-Q) plots for post-meta-analysis models for association between seasons of birth and DNA methylation (primary analyses) for at-birth (A and B) and childhood (C and D) samples. The Q–Q plots were generated by plotting observed p values (y-axis) against the expected uniform distribution of p values under the null hypothesis of no association (x-axis). Lambda and bias were estimated using BACON method [37]. All cohort-specific EWAS analyses were adjusted for gender of the child, gestational age at delivery, maternal age at delivery, maternal smoking during pregnancy, maternal socio-economic status, batch, child’s age at the time of sample collection and estimated cell proportions
Mean differences in methylation between autumn and each of the other two seasons were small in absolute terms [mean change in DNAm winter vs. autumn (cg26416241): 0.0034 (0.3%) and spring vs. autumn (cg18978324): 0.006 (0.6%)].
In the analysis of childhood samples (aged 1–11 years), differential DNAm was observed at four CpG sites (cg19416462, cg01656588, cg03263237 and cg15437053) in the summer-born children (FDR < 0.05, Table 1) when compared to the autumn-born children. One of the four CpGs, cg03263237, was a cross-reactive probe [46] and was excluded from further biological interpretations. The observed between-study heterogeneity was low to moderate (I2 < 50) for the FDR-significant CpGs. One of the four CpGs, cg19416462 (mapped to CD83), also passed the Bonferroni-corrected threshold p value. The remaining CpGs, after excluding the cross-reactive probe of the four childhood CpGs, two showed decreased methylation levels in children born in the summer compared to those born in the autumn in 92–100% of the cohorts, while cg15437053 was more methylated than the same CpG in autumn born children in 57% of the cohorts. The corresponding three CpGs in the at-birth samples of summer-born infants lacked even nominal significance but showed an overall tendency towards hypomethylation compared to the autumn-born infants (57–61% of the contributing cohorts). The mean differences in methylation (childhood analysis: summer vs. autumn) were small in absolute terms [mean change in DNAm (cg19416462): − 0.4%, (cg1656588): − 0.7%, (cg15437053): 0.4%].
Differentially methylated regions (DMRs) at birth and in childhood associated with season of birth
Several DMRs, containing at least two CpG sites, were associated with season of birth both in new-borns and in children aged 1–11 years (Table 2). There were 24 DMRs mapping to 27 known genes in the at-birth analysis. CpGs of the DMRs were mostly hyper-methylated, and their direction in the DMRs was the same in most cohorts (Additional file 5: Table S4). The DMRs identified and the genes mapped to these differential methylation signals were unique to the seasons apart from two DMRs mapped to HLA-F (in both winter and spring) and to AZU1 (in spring and summer, Table 2). The childhood analysis identified 32 DMRs mapped to 35 genes. CpGs of the DMRs were mostly hypo-methylated, and their direction in the DMRs was the same in most cohorts (Additional file 5: Table S4). Like in the at-birth analyses, the season of birth associated DMRs of childhood samples was unique to the season of birth except for two mapped genes, LOC441666 in the winter- and summer-born and HTR2A in winter- and spring-born children. There was no overlap between the at-birth and childhood DMRs.
Table 2.
Differentially methylated regions (DMRs) at birth and in childhood in different seasons
| Season of birtha | CHR | Region | CpGs in the region | Šidák p valueb | Minimum p valuec | Gene (hg19) | Gene group |
|---|---|---|---|---|---|---|---|
| At-birth DMRs | |||||||
| Winter | 7 | 1753371–1753934 | 7 | 1.7 × 10−6 | 9.4 × 10−7 | ELFN1 | intron, 5′UTR |
| 16 | 8806334–8807068 | 13 | 2.5 × 10−4 | 3.9 × 10−4 | ABAT | TSS, exon, 5′UTR, intron | |
| 1 | 59042906–59043601 | 10 | 6.9 × 10−4 | 3.6 × 10−4 | TACSTD2 | TSS, exon | |
| 6 | 29690797–29692424 | 17 | 1.5 × 10−3 | 3.6 × 10−4 | HLA-F | TSS, intron, 5′UTR, cds | |
| 11 | 2907309–2907955 | 9 | 3.0 × 10−3 | 1.0 × 10−3 | CDKN1C | Intergenic | |
| 7 | 1095391–1095745 | 6 | 4.1 × 10−3 | 1.0 × 10−3 | GPR146, C7orf50 | intron, 5′UTR, intron, intron, exon, 5′UTR | |
| 4 | 206087–206587 | 8 | 4.3 × 10−3 | 3.6 × 10−4 | ZNF876P | nc_intron, nc_exon | |
| 21 | 15134466–15135068 | 3 | 7.5 × 10−3 | 1.8 × 10−3 | LOC110091777 | nc_exon | |
| Spring | 11 | 368326–369217 | 24 | 2.9 × 10−8 | 2.6 × 10−9 | B4GALNT4 | intergenic |
| 6 | 31148307–31148773 | 16 | 3.2 × 10−7 | 1.4 × 10−7 | PSORS1C3 | nc_intron | |
| 6 | 29690741–29692607 | 27 | 5.8 × 10−6 | 6.4 × 10−7 | HLA-F | TSS, intron, 5′UTR, cds | |
| 6 | 41068528–41069073 | 8 | 5.9 × 10−6 | 5.8 × 10−7 | ADCY10P1, NFYA | nc_intron, nc_exon, exon, 3′UTR | |
| 12 | 132976697–132977165 | 4 | 1.2 × 10−4 | 2.4 × 10−5 | LOC101928416 | intergenic | |
| 17 | 17603506–17604209 | 6 | 1.6 × 10−4 | 3.9 × 10−5 | RAI1 | intron, 5′UTR | |
| 19 | 827690–828195 | 6 | 2.3 × 10−4 | 4.4 × 10−5 | AZU1 | TSS, intron, 5′UTR, cds | |
| 11 | 60534886–60535091 | 4 | 4.3 × 10−4 | 4.0 × 10−5 | MS4A15 | nc_intron, nc_exon, intron, cds, 5′UTR | |
| 19 | 11784489–11785150 | 10 | 4.7 × 10−4 | 8.4 × 10−7 | ZNF833P | nc_intron, nc_exon | |
| 12 | 6876163–6876346 | 2 | 1.3 × 10−3 | 9.3 × 10−5 | PTMS | Intron | |
| 11 | 1319623–1320659 | 6 | 1.2 × 10−3 | 1.3 × 10−2 | TOLLIP | intron, 5′UTR | |
| 20 | 35402098–35402548 | 13 | 4.2 × 10−3 | 2.5 × 10−4 | DSN1 | TSS, exon, 5′UTR | |
| 2 | 239139886–239140215 | 6 | 4.8 × 10−3 | 4.8 × 10−4 | LINC02610 | nc_intron, nc_exon | |
| 11 | 2721182–2721657 | 15 | 6.8 × 10−3 | 1. 8 × 10−4 | KCNQ1, KCNQ1OT1 | intron, nc_exon | |
| 11 | 47399788–47400355 | 9 | 7.6 × 10−3 | 8.1 × 10−4 | SPI1 | TSS, intron, 5′UTR, cds | |
| Summer | 19 | 827690–828195 | 6 | 3.8 × 10−4 | 1.1 × 10−3 | AZU1 | TSS, intron, 5′UTR, cds |
| 7 | 43288610–43288914 | 4 | 9.3 × 10−4 | 1.1 × 10−3 | HECW1 | Intron | |
| 13 | 20392381–20392731 | 5 | 8.5 × 10−3 | 4.7 × 10−3 | ZMYM5 | intergenic | |
| Childhood DMRs | |||||||
| Winter | 1 | 153599454–153600181 | 8 | 9.6 × 10−13 | 9.5 × 10−11 | S100A13 | TSS, exon, 5′UTR, introns |
| 19 | 55476640–55477835 | 7 | 3.8 × 10−8 | 1.2 × 10−3 | NLRP2 | TSS, intron, exon, 5′UTR, nc_intron, nc_exon | |
| 13 | 36871621–36872371 | 14 | 4.0 × 10−8 | 4.2 × 10−7 | CCDC169, CCDC169-SOHLH2 | TSS, intron, 5′UTR, cds, exon | |
| 5 | 191461–192128 | 9 | 2.6 × 10−5 | 1.5 × 10−5 | LRRC14B | TSS, 5′UTR, cds | |
| 11 | 67383352–67384065 | 8 | 9.0 × 10−5 | 4.9 × 10−5 | DOC2GP | Intergenic | |
| 16 | 31159533–31159945 | 5 | 3.5 × 10−4 | 1.1 × 10−4 | PRSS36 | intron, cds | |
| 10 | 42862851–42863533 | 6 | 4.6 × 10−4 | 1.8 × 10−3 | LOC441666 | nc_intron, nc_exon | |
| 13 | 47472025–47472454 | 12 | 1.7 × 10−3 | 3.7 × 10−4 | HTR2A | Intergenic | |
| 13 | 113540164–113540656 | 6 | 2.9 × 10−3 | 6.0 × 10−4 | ATP11A | exon, 3′UTR | |
| 10 | 43447046–43447643 | 3 | 6.6 × 10−3 | 1.2 × 10−3 | LINC01264 | Intergenic | |
| 1 | 1003101–1003554 | 4 | 6.7 × 10−3 | 1.0 × 10−3 | LOC105378948 | nc_intron, nc_exons | |
| 1 | 2121014–2121546 | 4 | 7.7 × 10−3 | 1.1 × 10−3 | FAAP20, LOC112268219 | nc_intron, nc_exon, intron, exon, 5′UTR, 3′UTR, cds | |
| Spring | 1 | 230415160–230415693 | 6 | 5.3 × 10−9 | 2.5 × 10−8 | GALNT2 | 3′UTR, cds |
| 17 | 33759459–33760318 | 11 | 1.6 × 10−7 | 1.0 × 10−5 | SLFN12 | TSS, exon, 5′UTR, intron | |
| 13 | 47472025–47472454 | 12 | 2.1 × 10−5 | 1.0 × 10−5 | HTR2A | Intergenic | |
| 3 | 193922012–193922718 | 7 | 6.9 × 10−5 | 3.7 × 10−5 | LINC02036 | nc_intron, nc_exon | |
| 6 | 163148828–163149478 | 6 | 2.5 × 10−4 | 1.0 × 10−5 | PACRG | intron, 5′UTR, cds, TSS | |
| 6 | 31543194–31543711 | 11 | 2.1 × 10−3 | 1.3 × 10−3 | TNF | TSS, intron, 5′UTR, cds | |
| 8 | 1094459–1094929 | 6 | 4.7 × 10−3 | 9.8 × 10−4 | DLGAP2 | Intron | |
| 11 | 10715150–10715385 | 6 | 4.9 × 10−3 | 2.2 × 10−3 | IRAG1 | TSS, exon, 5′UTR | |
| 19 | 39737743–39737886 | 3 | 8.6 × 10−3 | 6.7 × 10−4 | IFNL4 | intron, 3′UTR, cds, nc_intron, nc_exon | |
| 19 | 10735981–10736380 | 7 | 9.5 × 10−3 | 1.4 × 10−3 | SLC44A2 | TSS, intron, 5′UTR, cds | |
| Summer | 10 | 123355243–123356361 | 9 | 3.0 × 10−8 | 3.9 × 10−7 | FGFR2 | nc_intron, TSS, intron, exon, 5′UTR |
| 7 | 27183108–27184878 | 47 | 5.0 × 10−7 | 2.1 × 10−6 | HOXA-AS3, HOXA3, HOXA5 | nc_intron, TSS, 5′UTR, cds | |
| 6 | 41394131–41395119 | 5 | 1.1 × 10−5 | 1.2 × 10−5 | LINC01276 | intergenic | |
| 17 | 21280992–21281532 | 5 | 2.0 × 10−5 | 1.3 × 10−5 | KCNJ12 | intron, 5′UTR | |
| 10 | 131697055–131698005 | 7 | 8.1 × 10−4 | 2.4 × 10−4 | EBF3 | Intron | |
| 1 | 247171209–247171863 | 10 | 1.0 × 10−3 | 2.4 × 10−4 | ZNF695, ZNF670-ZNF695 | TSS, 5′UTR, cds, nc_intron, nc_exon | |
| 5 | 2334377–2334996 | 5 | 1.7 × 10−3 | 1.4 × 10−4 | LOC100506858 | intergenic | |
| 10 | 42862851–42863619 | 9 | 1.7 × 10−3 | 3.7 × 10−4 | LOC441666 | nc_intron, nc_exon | |
| 7 | 148768516–148769245 | 3 | 4.4 × 10−3 | 8.0 × 10−4 | ZNF786 | cds | |
| 17 | 48278753–48279290 | 9 | 9.3 × 10−3 | 1.1 × 10−3 | COL1A1 | TSS, intron, 5′UTR, cds | |
Differentially methylated regions (DMRs) were identified using comb-p. The inputs for comb-p were the annotated meta-analysed outputs of at-birth and childhood blood EWASs
CHR chromosomes, UTR untranslated region, TSS transcription start site, cds coding sequence
aReference season in EWAS of individual cohorts: autumn
bRegions with spatially adjusted and Šidák multiple testing corrected p value < 0.05
cP value for the most significant CpG in a DMR identified by comb-p
DNA methylation at birth and in childhood by latitude
We performed additional at-birth and childhood meta-analyses in subgroups stratified by latitude to check if the observed temporal associations varied with latitude. Cohorts, all from the northern hemisphere, were divided into two latitude groups—higher latitude (≥ 50°N, 9 cohorts, N = 5580, 59.3%) and the lower latitude [37.2–50°N (12 cohorts, N = 3823, 40.7%)].
In the at-birth analyses, we identified a single CpG, cg06251958, from the spring-born babies of the higher latitude subgroup (FDR-adjusted p value = 0.001, Table 3) and none from the winter- or summer-borns (all against autumn). This CpG (mapped to PSMC2) also passed the Bonferroni p value threshold (1.06 × 10−7). There were seven significant (FDR p value < 0.05) at-birth CpGs, six in the spring-born and one in the winter-born infants in the lower latitude subgroup analyses. One of these seven CpGs, cg23369114, was a cross-reactive probe [46]. There was no overlap in the FDR-adjusted differentially methylated CpGs between the seasons in each latitude subgroup nor between the two latitude subgroups (higher vs. lower) in any given birth season. The characteristics of cohorts from the higher and lower latitudes were similar apart from the latitude of the place of birth (Additional file 1: Table S1B). These results, therefore, suggest that the temporal associations between season of birth and DNAm were also latitude specific.
Table 3.
Association between season of birth and DNA methylation in at-birth and childhood stratified by latitude
| Season of birtha | CpG | Coeff | SE | p value | FDR p value | Lambdab | CHR | Position | Mapped gene | Directionc | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| At-birth | |||||||||||
| Higher latitude subgroup (≥ 50°N) | |||||||||||
| Spring | cg06251958 | 0.0095 | 0.0016 | 2.0 × 10−9 | 0.001 | 1.01 | 7 | 102989081 | PSMC2 | + + ? + + + − − + | 78 |
| Lower latitude subgroup (< 50°N) | |||||||||||
| Spring | cg01113988 | − 0.0082 | 0.0015 | 7.13 × 10−8 | 0.028 | 1.06 | 6 | 106035528 | PREP | − − − + − − − − − − − + | 29.4 |
| cg22168386 | − 0.0103 | 0.002 | 1.34 × 10−7 | 0.028 | 13 | 24205538 | TNFRSF19 | − − − − + + ? − + − − − | 40.1 | ||
| cg17462107 | 0.003 | 0.0006 | 2.21 × 10−7 | 0.028 | 5 | 66199867 | MAST4 | + − + + ? + + + + + + − | 14.9 | ||
| cg23369114d | − 0.0086 | 0.0017 | 3.83 × 10−7 | 0.031 | 19 | 14198698 | C19orf67 | − ? − − + − − ? − − − − | 21.2 | ||
| cg11520554 | 0.0022 | 0.0004 | 4.14 × 10−7 | 0.031 | 15 | 80216034 | C15orf37; ST20 | + + + + + + + − + + − − | 70.3 | ||
| cg20954180 | 0.0052 | 0.001 | 4.64 × 10−7 | 0.031 | 3 | 54606265 | CACNA2D3 | + + + + + + + + + + + − | 23.6 | ||
| Winter | cg01801443 | − 0.0045 | 0.0007 | 1.65 × 10−10 | 7.77 × 10−5 | 1.02 | 6 | 168629778 | LOC105378137 | + − − − + − ? − − − − ? − − | 40.5 |
| Childhood | |||||||||||
| Higher latitude subgroup (≥ 50°N) | |||||||||||
| Winter | cg22709217 | 0.0129 | 0.0024 | 6.97 × 10−8 | 0.021 | 1.12 | 22 | 50311962 | ALG12; CRELD2 | + + + + + + + + | 36.2 |
| cg18124955 | 0.0013 | 0.0002 | 1.09 × 10−7 | 0.021 | 2 | 173292329 | ITGA6 | + + + + + + + + | 0 | ||
Significant CpGs identified in meta-analyses of EWAS summary results from 9 (≥ 50°N, N = 5580) and 12 (< 50°N, N = 3823) cohorts for the at-birth latitude analyses. The p values for the CpGs that passed the Bonferroni-corrected significance threshold (1.06 × 10−7) are shown in bold. Cohort-specific EWAS models were adjusted for sex of the child, gestational age and maternal age at delivery, maternal smoking during pregnancy, maternal socio-economic status, batch, and cell type proportions
Coeff: regression coefficient (change in mean methylation compared to autumn reference); SE: standard error of the coefficient; Lambda: genomic inflation factor; CHR: chromosome; I2: a measure of between-study variability
aReference season: autumn
bEstimated using the BACON method of van Iterson et al. Ref [37]:
c + : hyper-methylation; –: hypo-methylation
dCross-reactive probe (Chen et al., Ref: [46])
The childhood analyses of the higher latitude cohorts revealed two FDR-significant (also Bonferroni-significant) CpGs in the winter-born children and none in the spring- and summer-born. Furthermore, there was no overlap in CpG methylation signals between the at-birth and childhood samples. We did not perform childhood DNAm analysis for the lower latitude subgroup due to its small sample size (N = 936, Fig. 1).
Differentially methylated regions (DMRs) at birth and in childhood by latitude
Interrogation of at-birth DMRs of the higher latitude subgroup (≥ 50°N) identified 18 DMRs mapped to 20 unique genes in the winter-born and 25 DMRs (23 mapped genes) in the summer-born infants (Table 4). In contrast, there were relatively fewer DMRs (n = 7) in the spring-born infants of the higher latitude cohorts. Most of the at-birth DMRs identified in the new-borns were unique to the season of birth (winter, spring, or summer) except for DMRs which mapped to ELFN1, SEMA5B and LOC154449 (identified in winter- and summer-born children) and AZU1 (in spring- and summer-born).
Table 4.
Season of birth associated differentially methylated regions (DMRs) at birth in cohorts from latitudes ≥ 50°N
| Season of birtha | CHR | Region (hg19) | CpGs in the region | Šidák p valueb | Minimum p valuec | Gene | Gene group |
|---|---|---|---|---|---|---|---|
| Winter | 1 | 203733946–203734584 | 6 | 7.2 × 10−8 | 1.7 × 10−7 | LAX1 | TSS, intron, 5′UTR, cds; exon |
| 6 | 170571118–170572041 | 10 | 1.1 × 10−5 | 1.1 × 10−6 | LOC154449 | nc_intron, nc_exon | |
| 7 | 1753371–1753934 | 7 | 1.4 × 10−5 | 9.0 × 10−6 | ELFN1 | intron, 5′UTR | |
| 3 | 122712113–122712742 | 4 | 3.7 × 10−5 | 2.1 × 10−5 | SEMA5B | TSS, exon, 5′UTR; intron, nc_intron | |
| 19 | 35629676–35630499 | 10 | 4.2 × 10−5 | 2.1 × 10−5 | FXYD1 | TSS, intron, exon, 5′UTR | |
| 17 | 72442903–72443426 | 6 | 1.1 × 10−4 | 3.5 × 10−5 | GPRC5C | intron, 3′UTR, cds | |
| 11 | 299365–300797 | 12 | 1.8 × 10−4 | 5.3 × 10−3 | IFITM5 | TSS, 5′UTR, cds | |
| 1 | 223316194–223317573 | 13 | 2.4 × 10−4 | 5.3 × 10−3 | TLR5 | TSS, intron, exon, 5′UTR | |
| 20 | 61583512–61584184 | 11 | 1.1 × 10−3 | 2.5 × 10−4 | SLC17A9 | TSS, 5′UTR, cds; exon | |
| 8 | 39171595–39172145 | 9 | 2.6 × 10−3 | 4.2 × 10−4 | ADAM5 | intergenic | |
| 14 | 24780142–24780951 | 11 | 3.3 × 10−3 | 2.1 × 10−4 | LTB4R2; CIDEB; LTB4R | 3′UTR, cds; TSS, exon, 5′UTR; exon | |
| 15 | 45021092–45021786 | 3 | 3.4 × 10−3 | 6.7 × 10−4 | LOC100419583 | nc_intron, nc_exon | |
| 14 | 96180294–96181069 | 11 | 4.5 × 10−3 | 6.7 × 10−4 | TCL1A | nc_exon; TSS, 5′UTR, cds | |
| 4 | 122853439–122854430 | 9 | 4.7 × 10−3 | 7.7 × 10−4 | TRPC3 | intron, cds; TSS, 5′UTR | |
| 21 | 15077071–15077799 | 3 | 5.5 × 10−3 | 8.1 × 10−4 | LOC110091777 | intergenic | |
| 3 | 122640753–122641341 | 6 | 8.3 × 10−3 | 1.1 × 10−3 | SEMA5B | nc_intron, nc_exon; intron, cds | |
| 20 | 42543009–42543803 | 9 | 9.4 × 10−3 | 7.3 × 10−4 | TOX2 | TSS, intron, 5′UTR, cds | |
| 3 | 46759071–46759723 | 9 | 0.00998 | 1.3 × 10−3 | PRSS50 | TSS, intron, 5′UTR, cds | |
| Spring | 6 | 28058690–28059233 | 10 | 1.2 × 10−5 | 3.0 × 10−5 | ZSCAN12P1 | nc_exon |
| 19 | 827690–828195 | 6 | 2.9 × 10−4 | 1.8 × 10−4 | AZU1 | TSS, intron, 5′UTR, cds | |
| 11 | 368326–368939 | 16 | 6.5 × 10−4 | 2.5 × 10−4 | B4GALNT4 | intergenic | |
| 6 | 24646448–24646807 | 7 | 1.3 × 10−3 | 3.9 × 10−4 | KIAA0319 | intergenic | |
| 6 | 29648136–29648926 | 22 | 1.5 × 10−3 | 2.5 × 10−4 | ZFP57 | intron, exon, 5′UTR | |
| 6 | 31148307–31148773 | 16 | 4.1 × 10−3 | 9.0 × 10−4 | PSORS1C3 | nc_intron | |
| 6 | 29893901–29894366 | 16 | 6.0 × 10−3 | 5.9 × 10−5 | HCG4B | nc_exon | |
| Summer | 7 | 1733107–1734350 | 9 | 4.0 × 10−6 | 8.9 × 10−5 | ELFN1 | intron, 5′UTR |
| 20 | 57425954–57427846 | 62 | 4.8 × 10−5 | 1.2 × 10−4 | GNAS, GNAS-AS1 | intron, 5′UTR, nc_exon, 3′UTR | |
| 7 | 50628943–50630108 | 7 | 1.3 × 10−4 | 2.0 × 10−4 | DDC | intron, 5′UTR | |
| 19 | 827404–828195 | 8 | 1.7 × 10−4 | 8.9 × 10−5 | AZU1 | TSS, intron, 5′UTR, cds | |
| 10 | 134664446–134665203 | 6 | 2.5 × 10−4 | 2.0 × 10−4 | CFAP46 | intron, cds | |
| 14 | 101290531–101293115 | 26 | 2.8 × 10−4 | 2.0 × 10−4 | MEG3 | nc_intron, nc_exon | |
| 12 | 132973282–132974687 | 8 | 3.5 × 10−4 | 2.0 × 10−4 | LOC101928416 | intergenic | |
| 6 | 32046638–32049288 | 15 | 3.7 × 10−4 | 3.0 × 10−3 | TNXB | intron, cds | |
| 12 | 133414230–133415291 | 6 | 5.1 × 10−4 | 3.7 × 10−3 | CHFR | exon, 3′UTR | |
| 6 | 28226860–28227507 | 11 | 1.3 × 10−3 | 3.8 × 10−4 | NKAPL; ZKSCAN4 | TSS, cds, intron, exon, 5′UTR | |
| 6 | 32051832–32054119 | 23 | 2.1 × 10−3 | 7.9 × 10−3 | TNXB | intron, cds | |
| 6 | 32039771–32041859 | 20 | 2.9 × 10−3 | 5.5 × 10−3 | TNXB | intron, cds | |
| 19 | 5048401–5049160 | 4 | 3.6 × 10−3 | 6.8 × 10−4 | KDM4B | intron | |
| 1 | 2387645–2388098 | 5 | 4.2 × 10−3 | 6.0 × 10−4 | PLCH2 | intergenic | |
| 7 | 24323236–24324460 | 10 | 4.3 × 10−3 | 7.3 × 10−4 | NPY | TSS, intron, 5′UTR | |
| 7 | 130131233–130132478 | 37 | 4.6 × 10−3 | 6.8 × 10−4 | MEST | TSS, intron, 5′UTR, cds, exon | |
| 7 | 1753371–1753934 | 7 | 5.0 × 10−3 | 6.8 × 10−4 | ELFN1 | intron, 5′UTR | |
| 2 | 27664992–27665736 | 11 | 5.6 × 10−3 | 7.1 × 10−4 | NRBP1, KRTCAP3 | exon, 3′UTR, TSS, intron, 5′UTR, cds | |
| 3 | 122712113–122712922 | 5 | 5.7 × 10−3 | 7.1 × 10−4 | SEMA5B | TSS, exon, 5′UTR; intron, nc_intron | |
| 12 | 53183483–53184111 | 6 | 5.9 × 10−3 | 5.1 × 10−4 | KRT3 | 3′UTR, cds | |
| 6 | 170571118–170572041 | 10 | 6.0 × 10−3 | 4.3 × 10−4 | LOC154449 | nc_intron, nc_exon | |
| 20 | 57414014–57415202 | 15 | 6.5 × 10−3 | 7.6 × 10−4 | GNAS-AS1, GNAS | nc_intron, TSS, 5′UTR, cds | |
| 10 | 35893599–35894455 | 9 | 6.6 × 10−3 | 3.6 × 10−4 | GJD4 | TSS, exon, 5′UTR | |
| 7 | 57484128–57484846 | 6 | 8.7 × 10−3 | 7.9 × 10−4 | MIR3147 | intergenic | |
| 6 | 32043004–32043764 | 5 | 8.8 × 10−3 | 4.1 × 10−4 | TNXB | intron |
Differentially methylated regions (DMRs) were identified using comb-p. The inputs for comb-p were the annotated meta-analysed outputs of at-birth EWASs
CHR chromosomes, UTR untranslated region, TSS transcription start site, cds coding sequence
aReference season in EWAS of individual cohorts: autumn
bRegions with Šidák p value < 0.05
cP value for the most significant CpG in a DMR identified by comb-p
Examination of the biological functions of the genes mapped to the at-birth higher latitude DMRs revealed that most of the genes have roles, amongst others, in the functions of the central nervous system (CNS) including psychological disorders, cancer or inflammation and immunity (see Additional file 6: Table S5, for a more detailed, but not exhaustive, list of genes associated with at-birth DMRs and their functions). The DMRs identified in the new-borns from the lower latitude (< 50°N) cohorts were also unique to seasons of birth (Additional file 7: Table S6). None of the DMRs identified in the higher latitude subgroup were present in the lower latitude samples (compare Table 4 and Additional 7: Table S6).
Interrogation of childhood DMRs of the higher latitude subgroup (≥ 50°N) identified 47 DMRs which were mapped to 54 genes (winter: 21 DMRs and 24 genes, spring: 16 DMRs and 18 genes, summer: 10 DMRs and 12 genes, Additional file 7: Table S7). Most of these DMRs were unique to the season of birth (winter, spring, or summer) except for a DMR that mapped to S100A13 identified in both winter- and spring-born children and a second DMR that mapped to MIR7159 identified in both winter- and summer-born children. Furthermore, there was little overlap between the DMRs identified in the at-birth (Table 4) and childhood analyses of the higher latitude subgroup (Additional file 7: Table S7).
Four at-birth DMRs of the summer-born babies from the higher latitude subset mapped to imprinted genes, GNAS/GNAS-AS1, MEG3 and MEST, with two of them mapping to the GNAS locus (GNAS and GNAS-AS1). These imprinted genes were specific for the summer-born babies and were not identified in the new-borns of spring or winter (Table 4). Whilst DMRs associated with GNAS/GNAS-AS1 and MEG3 were present only in the at-birth samples from the higher latitude subgroup, a DMR associated with the MEST gene (mesoderm-specific transcript) was present in both at-birth and in early childhood from the higher latitude subgroup (compare Table 4 with Additional file 7: Table S7.
Trait enrichment analysis
Trait Enrichment analysis was carried out on CpGs with unadjusted meta-analysed association p value < 1.0 × 105 using EWAS Atlas [42]. This analysis revealed that, apart from enrichment for traits related to cancers or CNS-associated disorders, the enriched traits are mutually exclusive between at-birth and childhood analyses (Additional file 9). The traits with higher odds ratios for enrichment in the at-birth analysis were vitamin B12 supplements, polycystic ovary syndrome, maternal stress, systemic lupus erythematosus, and Behcet’s disease. These traits were enriched in the at-birth but not in the childhood samples and were mostly in new-borns of mothers with the early months of their pregnancy in the months of autumn to winter, which may indicate influence of exposures in the earlier parts of foetal development.
A similar trend of mutual exclusivity of enriched traits was seen when the new-borns of higher (≥ 50°N) and lower (< 50°N) latitudes were compared. While the higher latitude new-borns alone had traits enriched in exposure to pollutants (particulate matter, nitric oxide, and maternal arsenic exposure) and folic acid supplement, the lower latitude offspring had unique enriched traits associated with response to different treatments, mixed connective tissue disease, asthma, puberty, and household socio-economic status. Parental arsenic exposure and autistic spectrum disorder were common in the new-borns and young children of higher latitude, although weakly enriched in the childhood analysis.
Metastable epialleles
We tested CpGs with unadjusted p value < 0.05 in meta-analyses for enrichment of metastable epialleles (loci whose epigenetic modifications are established during early embryonic development [47]). Our analyses found no evidence of enrichment of metastable epialleles.
Discussion
This study investigated the association of season of birth and DNAm at two time points (at-birth and in childhood), and in latitude subgroups, in multiple cohorts recruited from regions across the northern hemisphere. Season of birth can be a proxy for seasonal variations in the length of the day, temperature, availability of sunlight, exposure to UV light, pollen, nutrition, seasonal infectious agents, air pollution, C-section and many more. Here, we show evidence for the existence of season of birth specific associations with DNAm at-birth as well as in childhood. The differential methylation patterns in at-birth samples followed a season-dependent annual periodicity (temporal effect). In addition, we found suggestive evidence for latitude-specific fluctuations in DMRs (a spatial effect). To the best of our knowledge, our study is a first to demonstrate such season of birth effects on DNAm as well as the latitude specificity in these effects in a large study population with participants from diverse geographical locations.
Season of birth specific differential DNA methylation
Our primary analyses found epigenome-wide significant season of birth associations at two and four CpGs in the at-birth and childhood blood samples, respectively. One of the four CpGs in the childhood samples was found to be a cross-reactive probe [46] and should be interpreted with caution. The remaining differentially methylated CpGs were unique for the birth seasons in the at-birth and childhood samples. Furthermore, there was no overlap in the CpG methylation signals observed across the two time points investigated (at-birth and childhood). Several DMRs associated with the birth seasons were identified in the at-birth and childhood analyses. CpG sites that are close to each other are known to be co-methylated, and together these sites can function as a regional unit [48]. At the individual CpG level, the exposure-induced changes in methylation of these CpGs are often small and with weak statistical evidence. However, analysis of pre-defined regions as functional units, each containing several co-methylated CpGs, may provide more statistical power to detect the associations with methylation signals. Half of the DMRs identified in this study were in gene locations such as coding sequences, transcriptional start sites and/or untranslated 5′ or 3′ ends. The DMRs identified were in the vicinity of genes with known functions, amongst others, in tumorigenesis, psychiatric/neurological disorders or inflammation and immunity. Most DMRs and their mapped genes in the at-birth (24 out of 26) and childhood (30 out of 32) DMRs were specific to the birth seasons. The seasonal specificity of DMRs was consistent with the observed temporal effect of DNAm signals at the CpG level both at birth and in childhood. Like in the case of differentially methylated season-of-birth-associated CpGs, there was little overlap between most of the at-birth and childhood DMRs identified which may indicate that the at-birth methylation signals associated with season of birth did not persist over time.
Association between season of birth and DNA methylation is latitude dependent
Latitude-stratified analyses (higher latitude: ≥ 50°N and lower latitude: < 50°N) were carried out to explore the impact of latitude on seasonal differences in at-birth and childhood DNAm. One of the significant CpGs of the at-birth lower latitude analysis, cg23369114 (Table 3) was a cross-reactive probe identified by Chen et al. [46] which warrants caution while interpreting the results. Like in the primary analyses, temporal associations (season-specific) were observed in the two latitude subgroups for the at-birth and the higher latitude childhood analyses both at the CpG (6/7 CpGs) and DMR levels. Furthermore, the DMRs of at-birth and childhood analyses of the higher latitude subgroup were mutually exclusive except for two DMRs which were present at both time points. This is consistent with the findings in the primary analysis that majority of the at-birth associations between season of birth and methylation signals did not persist during childhood. Like in the primary analyses, the observed associations between season of birth and DNAm signals (CpG/DMR) were unique for the latitudes in each of the seasons. These findings point to a latitude-dependent spatial effect of the association between DNAm and season of birth.
The absence of sustained signals from birth to childhood in this study does not necessarily imply absence of persistence in general. There could be several reasons for this. The smaller sample size of the childhood analysis compared to that of the at-birth analysis might be one of the reasons. Choice of cord blood for the at-birth studies could have masked the birth to childhood persistence of DNAm signals in this study. For example, Paquette and Marsit highlighted the importance of placenta samples for studies on the influence of intrauterine HTR2A gene expression on early childhood and/or lifelong health outcomes [49]. The current study identified a DMR on chromosome 13 (47472025–47472454) in the winter-born children of the primary and the higher latitude subgroup childhood analyses. The same DMR, mapped to HTR2A (codes for serotonin receptor), was present in the spring-born children of the primary childhood analysis, but not in any of the at-birth analyses that were based mostly on cord blood data. Yet another factor could be the absence of paired comparisons in our study. The at-birth and childhood comparisons were done with > 80% of unpaired data as most cohorts contributed to either at-birth or childhood data. This meant that DNAm measurements at the two time points did not come from the same child. This might have made the true persistent signals, if there were any, undetectable. An analysis with paired data from the six cohorts (data available from the same child at both the time points) would have had limited power to detect methylation signals of small DMR sizes as such data were available for < 20% of the study population. The heterogeneity of the age group in the childhood analysis may also have contributed to the non-persistence of the at-birth DNAm signals. Age effects may be strong, and the age range was wide (1–11 years). It is likely that there may be some remaining effect despite controlling for child’s age in the models.
It may be noted that persistence of DNAm signals from birth to childhood is not necessarily needed for differential methylation to have an effect. Differential methylation at birth, which affects developmental programmes at a critical period, may have long-lasting effects, even if the differential methylation does not persist into early childhood.
The season-related DNAm signals (CpGs and DMRs) identified in the primary and latitude analyses were non-overlapping. The primary analysis investigated the seasonal variations disregarding the information on the latitude of birth of the children. The results make use of the distribution of methylation signals across the seasons only. In contrast, the latitude analysis is a stratified analysis which examines the above seasonal variations in two separate latitude regions (lower and higher latitudes in the northern hemisphere). The results of such an analysis look for differences in the distribution of methylation signals across the seasons in two separate latitude subgroups which are likely to be different from that of the primary analysis. However, the DMR mapped to HTR2A was present both in the winter- and spring-born children of the preliminary childhood analysis as well as the winter-born children of the higher latitude childhood analysis. Alternatively, the non-overlapping methylation signals observed in the latitude subgroups, despite smaller sample sizes and stringent FDR cut offs, are likely to be the enriched latitude-specific signals which were not visible in the primary analysis.
Genes implicated in the season of birth association of DNA methylation
Several of the differentially methylated probes (DMPs) and DMRs identified in our analyses mapped to genes with well characterised functions. We highlight below some of the genes mapped to DMPs or DMRs identified in the at-birth and childhood analyses of the higher latitude subgroup which are likely to be associated with outcomes relevant to season of birth.
The most studied link between season of birth and a disease outcome is that of schizophrenia (SZ). In the general population in the northern hemisphere, individuals born in late winter and early spring have an increased risk of developing SZ later in life [9, 50, 51]. In addition, the prevalence of SZ has also been shown to increase with increasing latitude with the highest prevalence rates at the poles [52, 53]. Correlation patterns of risk factors and SZ identified pre-natal vitamin D deficiency and infections like influenza and toxoplasmosis which are more prevalent in higher latitude and cold climates, as major risk factors of SZ. Interestingly, DNAm of cg12022621, mapped to the LAX1 gene (Lymphocyte Transmembrane Adaptor 1), has been shown to be associated with severity of certain symptoms of schizophrenia (SZ) in a case–control study [54]. LAX1 is also the mapped gene for one of the top-ranked at-birth DMRs (Šidák-corrected p value: 7.2 × 10−8) spanning six CpGs including cg12022621 (crude p value: 3 × 10−4, Table 4) and was identified only in the winter-born group of the higher latitude subset in the at-birth samples. While LAX1-associated SZ was absent from the childhood DMR analyses (Table 4), published literature reveals SZ to be one of the most highlighted outcomes of the major depressive disorder (MDD) family of mental health outcomes in winter- and spring-born children (Additional file 6: Table S5 and references therein). The corresponding associated genes in the childhood analyses were LY6G5C, HTR2A, SHANK 1 and NKAPL, in the winter-born children and, SNTG2 in the spring-borns [55–59].
Other genes associated with neurocognitive disorders, neural development, or the central nervous system, not exclusively, in the at-birth analyses of the higher latitude include ELF1N, SEMA5B, GPRC5C, SLC17A9, TOX2, PRSS50, AZU1, KIAA0319, PSORS1C3, GNAS, GNAS-AS1, DDC, CFAP46, MEG3, TNXB, ZKSCAN4, PLCH2, NPY, NRBP1, LTB4R and LTB4R2 in the winter-born babies (Additional file 6: Table S5 and references therein). The same in the childhood analyses (higher latitude) was S100A13, MIR-647, ZKSCAN4, NAV2, NCK2 and CAT in the winter-born children, S100A13, SZT2, C11orf21, PACRG, DLEU7 and TAPBP in the spring-borns, and FARS2, CHKB and RPH3AL in the summer-born children (see references in Additional file 8: Table S8).
Of note, SLC17A9, mapped by a DMR unique to the at-birth analyses of infants of the higher latitude subgroup born in winter (Additional file 6: Table S5), is associated with a skin-specific autoinflammatory disease, disseminated superficial actinic porokeratosis (DSAP) [60]. Induction and exacerbation of DSAP are known to be linked to exposure to sunlight or artificial ultraviolet radiation [61]. Similarly, other genes linked to skin disorders include PSORS1C3 (spring-born), and LTB4R (winter-born), both of which are known to be linked to the autoimmune disease psoriasis [62–64]. Interestingly, prevalence of psoriasis is higher in populations of higher latitude regions of northern Europe [65]. The DMR-associated genes responsible for skin disorders in the winter-born children of higher latitude subgroup were different from those in the at-birth samples. The childhood genes were MIR-1914, MIR-647 and MAP3K8 (common wart) and TNF (psoriasis) (Additional file 8: Table S8) [66–68].
A DMR on chromosome 10, identified in the summer-born children of the higher latitude subgroup childhood analysis, was mapped to FGFR2 gene. Interestingly, alterations in the FGFR2 gene seen in several cancer types made it a target for the development of person-specific treatments [69–72]. DNAm status in the proximity of the FGFR2 gene, such as the one observed in this study, may further influence the FGFR2 alterations in the cancer patients. Therefore, knowledge of methylation status around FGFR2 of the cancer patients will help fine tune the design of the person-specific therapies further.
Several genes mapped to DMRs of at-birth blood samples of the winter-born of the higher latitude, not found in the lower latitude, had links to immunity and inflammation (LAX1: T-/B-/NK-cell activation; TLR5: pathogen recognition; LTB4R: pathogenesis of inflammatory diseases; PSORS1C3: psoriasis; TCL1A: modulation of immune responses; ADAM5: autoimmune diseases) [54, 62–64, 73–76]. LTB4R, leukotriene B4 receptor, appears to be the most epigenetically divergent gene in the peripheral blood of humans. LTB4R has been linked to a variety of inflammatory diseases such as asthma [77], allergic airway inflammation [78], inflammatory arthritis [79], atherosclerosis [80], inflammatory bowel disease [81] and psoriasis [82]. This is significant as the pregnant mothers of the winter-born babies would have been exposed to various seasonal factors that are responsible for allergy/asthma and other causal exposures of inflammation during the first and the second trimesters of their pregnancy. Lockett et al. hypothesised that the season-associated DNAm of allergic diseases such as eczema most probably arose postnatally since no such association was observed at birth (in cord blood) in their study [26]. However, the lack of evidence for a prenatal association at an epigenome-wide level could also have been due to the modest sample size of their study (N = 175).
A CpG, cg003488551, mapped to C7orf50 and DMR containing this CpG were reported to be associated with prenatal exposure to particulate air pollution in a meta-analysis by Gruzieva et al. [83]. Major air pollutants show seasonal patterns with highest concentrations in the indoor heating seasons of November to February (winter months) in the northern hemisphere [84]. It is possible the summer-born babies in the current study were exposed to indoor pollutants during most part of their in utero life. However, our study did not identify the same methylation signals in babies born in the higher latitude subgroup with extended and extreme winter conditions. This study also did not identify any known vitamin D-associated CpGs or genes mapped to DMRs in cohorts from the higher latitude (≥ 50°N).
Imprinted genes and metastable epialleles
Our study also identified DMRs associated with three imprinted genes in the at-birth analyses of the summer-born infants of the higher latitude, GNAS/GNAS-AS1, MEG3 and MEST (PEG1). Almost all the CpGs found in the DMRs of these imprinted genes were hyper-methylated. In addition to being season of birth and latitude specific (only in the summer-born babies of the higher latitude), they were not found in the older children from the higher latitude subgroup. However, a DMR associated with the MEST gene (mesoderm-specific transcript) was present both at birth (summer-born) and in early childhood (spring-born) from the higher latitude subgroup. Many imprinted genes function as regulators of embryonic or neonatal growth and may therefore influence a spectrum of heritable outcomes later in life. The majority of imprinted genes are expressed in the brain, and methylation of these genes in their imprinting control regions (ICRs) has been implicated in neuropsychiatric disorders (reviewed in [85]). For example, GNAS is a complex imprinted locus with five gene products and multiple DMRs in four of these genes. These DMRs are shown to be associated with a genetic disorder known as pseudohypoparathyroidism type-Ib (PHP-Ib) (reviewed in [86]). Furthermore, a UK Biobank GWAS study of 113,000 individuals with insomnia identified GNAS as a potential gene candidate in females [87]. A previous study reported that the retention of a sex-specific association between a hyper-methylated DMR associated with MEST and weight status from birth to early childhood [88].
Metastable epialleles are variably methylated loci with cross-tissue methylation signatures indicative of establishment in the early embryo [44]. They, therefore, provide a useful tool for examining the timing of exposure driven DNAm changes in easily accessible tissues such as blood that may serve as a proxy for patterns of systemic methylation. Silver et al. demonstrated elevated DNAm levels at putative metastable epialleles in rural Gambian children who were conceived during the rainy season compared to those conceived in the dry season [47]. We checked for the enrichment of metastable epialleles in each of the seasons of birth, but none were found, providing no evidence for a season of conception effect at these loci.
Implications of temporal and latitude-dependent associations between season of birth and differential DNA methylation
In the at-birth meta-analyses, there were only a few robust but weak genome-wide associations between CpG methylation and season of birth. DMR analysis is believed to be statistically more powerful than the analysis of individual probes as it combines methylation signals from nearby CpGs to give more reliable signals for associations between DNAm and exposures. It is possible that the seasonal variations in exposures influence DNAm via groups of CpG sites over an extended region as in the DMRs where contiguous differential methylation may be maintained. Our findings on the birth season-dependent variations of regional epigenetic signals concur with the findings of a study by Dopico et al. which demonstrated the existence of seasonal variations in gene expression profiles over a year in ethnically and geographically diverse populations [21]. They attributed these seasonal variations in gene expression profiles to the seasonal changes in the cellular composition of blood. However, unlike in the study by Dopico et al., our models for association between birth season and DNAm were adjusted for cellular heterogeneity and therefore, the findings are unlikely to be the result of seasonality of blood composition. Epigenetic marks including DNAm signals, whether acting at the individual CpG site or DMR level, are dynamic and may not originate stochastically. Oh & Petronis and Oh et al. proposed that DNAm variability occurring in a periodic fashion over twelve months (circannual oscillations) could contribute to variations in the severity of seasonal diseases [89, 90]. We conclude that our findings on seasonal variations in DNAm signals including DMRs are in line with the circannual oscillations proposed by Oh & Petronis and are likely to reflect the influence of seasonal exposures on later life health events.
Strengths and Limitations
Our study investigated associations between season of birth and DNAm on a large scale with 9358 at-birth and 3610 childhood samples. Analyses with such a large sample size, especially for the at-birth samples, make it possible to interpret results with more confidence. The cohort-specific EWAS analyses were robust and adjusted for several pre-specified potential confounders, including cell type proportions. The cohorts originated from geographically diverse locations in terms of latitude in the northern hemisphere: 32.7–71.2°N and 36.7–58.7°N for the at-birth and childhood samples. This enabled us to carry out latitude stratifications to investigate season of birth associations of DNAm. Of the 27 cohorts analysed in this study, more than 95% of the participants were of European ancestry. It remains to be seen whether our results are generalizable to other populations.
This study has limitations. This study was designed only to examine the associations between DNAm and season of birth as an exposure and not the factors for which season of birth is a proxy (see above). Heterogeneity of the age range in the childhood population in this study and its smaller sample size (childhood: N = 3569 vs. at-birth: N = 9358) made it harder to interpret with certainty the absence of at-birth DNAm signals in childhood data. Another limitation of our study is that the at-birth and childhood data were not from the same children for most of the cohorts (21/27 studies). A longitudinal study which follows up the same children at different latitudes in sufficient numbers is necessary to detect the persistence of at-birth DNAm signals from birth to childhood. The latitude analyses of this study have not been adjusted for longitude. The same latitude can have geographic locations that have very different climates, e.g. New York and Madrid (40°N). Furthermore, the choice of autumn as a reference season may have masked some of the other significant associations between season of birth and DNAm. This study was initiated as a follow-on from an earlier study on seasonality effects on asthma and allergy outcomes which showed the strongest effects on allergy phenotypes with autumn as the reference season [26]. However, we expect to observe the circannual oscillations in DNAm even if a season other than autumn was used as the reference season. The cohorts which contributed the EWAS summary results pre-processed and analysed their data using their preferred pipelines and this may have influenced our results. However, Joubert et al. found that their results were robust to different normalization methods used across studies and cell type adjustment [24]. Furthermore, Lussier et al. demonstrated that while different pipelines give different EWAS associations at a set significance threshold their magnitude and directions were consistent [91].
Conclusions
In this large epigenome-wide meta-analysis study, we provide evidence for an association between season of birth and differential DNAm that is unique for the seasons of the year (temporal effect) and suggestive evidence of a latitude (spatial) effect. Findings in this study add to the understanding of a potential epigenetic role in the seasonality of human disease. Our study suggests the existence of a circannual periodicity in DNAm patterns, much like the seasonal periodicity observed in gene expression profiles.
Supplementary Information
Additional file 1. Table S1: “Summary of the studies in the meta-analysis for the association between Season of Birth and DNA methylation at birth and in children (age: 1 to 11 years)”. Baseline summary of participants from each of the individual cohorts.
Additional file 2. “Cohort-specific methods and declarations (cohorts listed in alphabetical order)”. Method description, Funding and Acknowledgements from the participating cohorts.
Additional file 3. “PACE analysis plan for Season of Birth and methylation profiles in children (6 June 2018)”. Analysis plan that was circulated amongst the participant cohorts for the Season of Birth study.
Additional file 4. Table S2: “Models used in this meta-analysis study”. Inflation (lambda) and bias information of the models. Table S3: “Comparison of magnitude and direction of the FDR-significant DNA methylation signals identified in the at-birth and childhood meta-analyses”.
Additional file 5. Table S4: “Direction of differential methylation of CpGs in DMRs of the at-birth and childhood analyses (preliminary analysis”).
Additional file 6. Table S5: “Genes mapped to DMP/DMR identified in at-birth samples of babies born in the latitude ≥ 50°N and some examples of their associations with biological functions”. Provides examples of known functional associations of genes mapped to significant CpG sites or differentially methylated regions identified in this study.
Additional file 7. Table S6: “Season of birth-associated differentially methylated regions (DMRs) at birth in babies born in latitudes < 50°N”. Differentially methylated regions and mapped genes identified in the at-birth samples of babies born in latitudes < 50°N (lower latitude subgroup analysis). Table S7: “Season of birth associated with differentially methylated regions (DMRs) in children born in latitudes ≥ 50°N”.
Additional file 8. Table S8: “Genes mapped to DMR identified in the childhood samples of children born in the latitude ≥ 50°N and some examples of their associations with biological functions”. Provides examples of known functional associations of genes mapped to significant differentially methylated regions identified in this study.
Additional file 9. “Trait Enrichment Analysis (EWAS Atlas)”. Trait names and the odds ratios for their association with CpG sites for all the models meta-analysed in this study.
Abbreviations
- DMR
Differentially methylated region
- DNAm
DNA methylation
- EWAS
Epigenome-wide association study
- FDR
False discovery rate
- PACE
Pregnancy And Childhood Epigenetics
- SNP
Single nucleotide polymorphism
- SES
Socio-economic status
- SZ
Schizophrenia
Author contributions
CHW, FR, JWH conceived, designed the study, and formulated the analysis strategy; LK, MZA, FIR carried out the data curation, formal analysis, methodology and visualization; LK drafted the manuscript and prepared Tables and figures; LK, CHW, MZA, AG, MJS, JWH revised the manuscript critically and extensively. Cohort-specific authors performed and/or supervised the EWAS analyses that were meta-analysed. All authors read, reviewed, and approved the manuscript.
Funding
See Additional file 2 for funding details.
Availability of data and materials
Individual cohort-level data can be obtained from the respective cohort (see Additional file 1: Table S1 and Additional file 2 for cohort details).
Declarations
Ethics approval and consent to participate
All studies included in this meta-analysis obtained ethical approval from the relevant committees and informed consent from study participants or the parents/legal guardian if a child participant (see Additional file 2 for details).
Competing interests
None declared any competing interests.
Footnotes
Catarina Almqvist onwards: Senior co-authors listed in reverse alphabetical order of the cohort names
Faisal I. Rezwan and John W Holloway: Joint senior authors
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Esther Walton to Emily Oken: Junior co-authors listed according to cohort names ordered in alphabetical order
References
- 1.Weber GW, Prossinger H, Seidler H. Height depends on month of birth. Nature. 1998;391(6669):754–755. doi: 10.1038/35781. [DOI] [PubMed] [Google Scholar]
- 2.Brennan PJ, Greenberg G, Miall WE, Thompson SG. Seasonal variation in arterial blood pressure. Br Med J Clin Res Ed. 1982;285(6346):919–923. doi: 10.1136/bmj.285.6346.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon DJ, Trost DC, Hyde J, et al. Seasonal cholesterol cycles: the Lipid Research Clinics Coronary Primary Prevention Trial placebo group. Circulation. 1987;76(6):1224–1231. doi: 10.1161/01.cir.76.6.1224. [DOI] [PubMed] [Google Scholar]
- 4.Foster RG, Roenneberg T. Human responses to the geophysical daily, annual and lunar cycles. Curr Biol. 2008;18(17):R784–R794. doi: 10.1016/j.cub.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Doblhammer G, Vaupel JW. Lifespan depends on month of birth. Proc Natl Acad Sci USA. 2001;98(5):2934–2939. doi: 10.1073/pnas.041431898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pell JP, Cobbe SM. Seasonal variations in coronary heart disease. QJM. 1999;92(12):689–696. doi: 10.1093/qjmed/92.12.689. [DOI] [PubMed] [Google Scholar]
- 7.Iikuni N, Nakajima A, Inoue E, et al. What’s in season for rheumatoid arthritis patients? Seasonal fluctuations in disease activity. Rheumatology. 2007;46(5):846–848. doi: 10.1093/rheumatology/kel414. [DOI] [PubMed] [Google Scholar]
- 8.Moltchanova EV, Schreier N, Lammi N, Karvonen M. Seasonal variation of diagnosis of Type 1 diabetes mellitus in children worldwide. Diabet Med. 2009;26(7):673–678. doi: 10.1111/j.1464-5491.2009.02743.x. [DOI] [PubMed] [Google Scholar]
- 9.Cheng C, Loh E-W, Lin C-H, Chan C-H, Lan T-H. Birth seasonality in schizophrenia: effects of gender and income status. Psychiatry Clin Neurosci. 2013;67(6):426–433. doi: 10.1111/pcn.12076. [DOI] [PubMed] [Google Scholar]
- 10.Koskinen A, Lemmetyinen R, Luukkainen A, et al. Season of birth affects the risk of adult-onset asthma in Finland. Allergy. 2023;78(2):555–558. doi: 10.1111/all.15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudsen TB, Thomsen SF, Ulrik CS, Fenger M, Nepper-Christensen S, Backer V. Season of birth and risk of atopic disease among children and adolescents. J Asthma. 2007;44(4):257–260. doi: 10.1080/02770900701246832. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson L, Björkstén B, Hattevig G, Kjellman B, Sigurs N, Kjellman NI. Season of birth as predictor of atopic manifestations. Arch Dis Child. 1997;76(4):341–344. doi: 10.1136/adc.76.4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keet CA, Matsui EC, Savage JH, et al. Potential mechanisms for the association between fall birth and food allergy. Allergy. 2012;67(6):775–782. doi: 10.1111/j.1398-9995.2012.02823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thysen AH, Rasmussen MA, Kreiner-Møller E, et al. Season of birth shapes neonatal immune function. J Allergy Clin Immunol. 2016;137(4):1238–1246.e13. doi: 10.1016/j.jaci.2015.08.041. [DOI] [PubMed] [Google Scholar]
- 15.Torkildsen O, Grytten N, Aarseth J, Myhr K-M, Kampman MT. Month of birth as a risk factor for multiple sclerosis: an update. Acta Neurol Scand Suppl. 2012;195:58–62. doi: 10.1111/ane.12040. [DOI] [PubMed] [Google Scholar]
- 16.Hsu C-W, Tseng P-T, Tu Y-K, et al. Month of birth and mental disorders: a population-based study and validation using global meta-analysis. Acta Psychiatr Scand. 2021;144(2):153–167. doi: 10.1111/acps.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaiserman A. Season-of-birth phenomenon in health and longevity: epidemiologic evidence and mechanistic considerations. J Dev Orig Health Dis. 2021;12(6):849–858. doi: 10.1017/S2040174420001221. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Nishio H, Tarutani Y, et al. Seasonal stability and dynamics of DNA methylation in plants in a natural environment. Genes. 2019;10(7):E544. doi: 10.3390/genes10070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viitaniemi HM, Verhagen I, Visser ME, Honkela A, van Oers K, Husby A. Seasonal variation in genome-wide DNA methylation patterns and the onset of seasonal timing of reproduction in great tits. Genome Biol Evol. 2019;11(3):970–983. doi: 10.1093/gbe/evz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson TJ, Prendergast BJ. Reversible DNA methylation regulates seasonal photoperiodic time measurement. Proc Natl Acad Sci USA. 2013;110(41):16651–16656. doi: 10.1073/pnas.1310643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dopico XC, Evangelou M, Ferreira RC, et al. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat Commun. 2015;6:7000. doi: 10.1038/ncomms8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricceri F, Trevisan M, Fiano V, et al. Seasonality modifies methylation profiles in healthy people. PLoS ONE. 2014;9(9):e106846. doi: 10.1371/journal.pone.0106846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leenen FAD, Muller CP, Turner JD. DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenet. 2016;8:92. doi: 10.1186/s13148-016-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98(4):680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiklund P, Karhunen V, Richmond RC, et al. DNA methylation links prenatal smoking exposure to later life health outcomes in offspring. Clin Epigenet. 2019;11(1):97. doi: 10.1186/s13148-019-0683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lockett GA, Soto-Ramírez N, Ray MA, et al. Association of season of birth with DNA methylation and allergic disease. Allergy. 2016;71(9):1314–1324. doi: 10.1111/all.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix JF, Joubert BR, Baccarelli AA, et al. Cohort profile: Pregnancy And Childhood Epigenetics (PACE) consortium. Int J Epidemiol. 2018;47(1):22–23u. doi: 10.1093/ije/dyx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markunas CA, Wilcox AJ, Xu Z, et al. Maternal age at delivery is associated with an epigenetic signature in both newborns and adults. PLoS ONE. 2016;11(7):e0156361. doi: 10.1371/journal.pone.0156361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosin-Tomas M, Cilleros-Portet A, Aguilar-Lacasaña S, Fernandez-Jimenez N, Bustamante M. Prenatal maternal smoke, DNA methylation, and multi-omics of tissues and child health. Curr Environ Health Rep. 2022;9(3):502–512. doi: 10.1007/s40572-022-00361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Govender P, Ghai M, Okpeku M. Sex-specific DNA methylation: impact on human health and development. Mol Genet Genomics. 2022;297(6):1451–1466. doi: 10.1007/s00438-022-01935-w. [DOI] [PubMed] [Google Scholar]
- 31.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hattab MW, Shabalin AA, Clark SL, et al. Correcting for cell-type effects in DNA methylation studies: reference-based method outperforms latent variable approaches in empirical studies. Genome Biol. 2017;18(1):24. doi: 10.1186/s13059-017-1148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomon O, MacIsaac J, Quach H, et al. Comparison of DNA methylation measured by Illumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. Epigenetics. 2018;13(6):655–664. doi: 10.1080/15592294.2018.1497386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Iterson M, van Zwet EW, BIOS Consortium, Heijmans BT. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017;18(1):19. [DOI] [PMC free article] [PubMed]
- 38.Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 40.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28(22):2986–2988. doi: 10.1093/bioinformatics/bts545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reese SE, Xu C-J, den Dekker HT, et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. J Allergy Clin Immunol. 2019;143(6):2062–2074. doi: 10.1016/j.jaci.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Zou D, Li Z, et al. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019;47(D1):D983–D988. doi: 10.1093/nar/gky1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silver MJ, Kessler NJ, Hennig BJ, et al. Independent genomewide screens identify the tumor suppressor VTRNA2-1 as a human epiallele responsive to periconceptional environment. Genome Biol. 2015;16:118. doi: 10.1186/s13059-015-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kessler NJ, Waterland RA, Prentice AM, Silver MJ. Establishment of environmentally sensitive DNA methylation states in the very early human embryo. Sci Adv. 2018;4(7):eaat2624. [DOI] [PMC free article] [PubMed]
- 45.Van Baak TE, Coarfa C, Dugué P-A, et al. Epigenetic supersimilarity of monozygotic twin pairs. Genome Biol. 2018;19(1):2. doi: 10.1186/s13059-017-1374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silver MJ, Saffari A, Kessler NJ, et al. Environmentally sensitive hotspots in the methylome of the early human embryo. Elife. 2022;11:e72031. doi: 10.7554/eLife.72031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD, Kahraman A, Law CW, et al. Statistical methods for detecting differentially methylated loci and regions. Front Genet. 2014;5:324. doi: 10.3389/fgene.2014.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paquette AG, Marsit CJ. The developmental basis of epigenetic regulation of HTR2A and psychiatric outcomes. J Cell Biochem. 2014;115(12):2065–2072. doi: 10.1002/jcb.24883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torrey EF. Prevalence studies in schizophrenia. Br J Psychiatry. 1987;150:598–608. doi: 10.1192/bjp.150.5.598. [DOI] [PubMed] [Google Scholar]
- 51.Mortensen PB, Pedersen CB, Westergaard T, et al. Effects of family history and place and season of birth on the risk of schizophrenia. N Engl J Med. 1999;340(8):603–608. doi: 10.1056/NEJM199902253400803. [DOI] [PubMed] [Google Scholar]
- 52.Davies G, Welham J, Chant D, Torrey EF, McGrath J. A systematic review and meta-analysis of Northern Hemisphere season of birth studies in schizophrenia. Schizophr Bull. 2003;29(3):587–593. doi: 10.1093/oxfordjournals.schbul.a007030. [DOI] [PubMed] [Google Scholar]
- 53.Kinney DK, Teixeira P, Hsu D, et al. Relation of schizophrenia prevalence to latitude, climate, fish consumption, infant mortality, and skin color: a role for prenatal vitamin d deficiency and infections? Schizophr Bull. 2009;35(3):582–595. doi: 10.1093/schbul/sbp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Chen J, Ehrlich S, et al. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophr Bull. 2014;40(4):769–776. doi: 10.1093/schbul/sbt080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wockner LF, Morris CP, Noble EP, et al. Brain-specific epigenetic markers of schizophrenia. Transl Psychiatry. 2015;5(11):e680. doi: 10.1038/tp.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheah S-Y, Lawford BR, Young RM, Morris CP, Voisey J. mRNA expression and DNA methylation analysis of serotonin receptor 2A (HTR2A) in the human schizophrenic brain. Genes. 2017;8(1):14. doi: 10.3390/genes8010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du J, Nakachi Y, Kiyono T, et al. Comprehensive DNA methylation analysis of human neuroblastoma cells treated with haloperidol and risperidone. Front Mol Neurosci. 2021;14:792874. doi: 10.3389/fnmol.2021.792874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prata DP, Costa-Neves B, Cosme G, Vassos E. Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: a systematic review. J Psychiatr Res. 2019;114:178–207. doi: 10.1016/j.jpsychires.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Sabaie H, Mazaheri Moghaddam M, Mazaheri Moghaddam M, et al. Long non-coding RNA-associated competing endogenous RNA axes in the olfactory epithelium in schizophrenia: a bioinformatics analysis. Sci Rep. 2021;11(1):24497. doi: 10.1038/s41598-021-04326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui H, Li L, Wang W, et al. Exome sequencing identifies SLC17A9 pathogenic gene in two Chinese pedigrees with disseminated superficial actinic porokeratosis. J Med Genet. 2014;51(10):699–704. doi: 10.1136/jmedgenet-2014-102486. [DOI] [PubMed] [Google Scholar]
- 61.Chernosky ME, Anderson DE. Disseminated superficial actinic porokeratosis. Clinical studies and experimental production of lesions. Arch Dermatol. 1969;99(4):401–407. doi: 10.1001/archderm.99.4.401. [DOI] [PubMed] [Google Scholar]
- 62.Iversen L, Kragballe K, Ziboh VA. Significance of leukotriene-A4 hydrolase in the pathogenesis of psoriasis. Skin Pharmacol. 1997;10(4):169–177. doi: 10.1159/000211501. [DOI] [PubMed] [Google Scholar]
- 63.Kato K, Yokomizo T, Izumi T, Shimizu T. Cell-specific transcriptional regulation of human leukotriene B(4) receptor gene. J Exp Med. 2000;192(3):413–420. doi: 10.1084/jem.192.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiśniewski A, Matusiak Ł, Szczerkowska-Dobosz A, Nowak I, Kuśnierczyk P. HLA-C*06:02-independent, gender-related association of PSORS1C3 and PSORS1C1/CDSN single-nucleotide polymorphisms with risk and severity of psoriasis. Mol Genet Genomics. 2018;293(4):957–966. doi: 10.1007/s00438-018-1435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrees V, Wolf S, Augustin M, Mohr N, Augustin J. Regional variations and prevalence of psoriasis in Germany from 2010 to 2017: a cross-sectional, spatio-epidemiological study on ambulatory claims data. BMJ Open. 2021;11(11):e047806. doi: 10.1136/bmjopen-2020-047806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Eitan LN, Alghamdi MA, Tarkhan AH, Al-Qarqaz FA. Gene expression profiling of MicroRNAs in HPV-induced warts and normal skin. Biomolecules. 2019;9(12):757. doi: 10.3390/biom9120757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alghamdi MA, Al-Eitan LN, Tarkhan AH, Al-Qarqaz FA. Global gene methylation profiling of common warts caused by human papillomaviruses infection. Saudi J Biol Sci. 2021;28(1):612–622. doi: 10.1016/j.sjbs.2020.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belinchon Romero I, Mateu Puchades A, Ribera Pibernat M, Ruiz Genao DianaP, de la Cueva Dobao P, Carrascosa JM. Criteria used to define tumor necrosis factor-alpha inhibitors failure in patients with moderate-to-severe psoriasis: a systematic literature review. Ann Med. 2022;55(1):1335–45. [DOI] [PMC free article] [PubMed]
- 69.Ilyas SI, Affo S, Goyal L, et al. Cholangiocarcinoma—novel biological insights and therapeutic strategies. Nat Rev Clin Oncol. 2023;6:66. doi: 10.1038/s41571-023-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palmieri L-J, Soubeyran I, Pernot S. Oesogastric cancer—new therapeutic targets. Bull Cancer. 2023;110(5):560–569. doi: 10.1016/j.bulcan.2022.08.005. [DOI] [PubMed] [Google Scholar]
- 71.Zeng W, Mao R, Zhang Z, Chen X. Combination therapies for advanced biliary tract cancer. J Clin Transl Hepatol. 2023;11(2):490–501. doi: 10.14218/JCTH.2022.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Shi T, Wang X, et al. FGFR2 alteration as a potential therapeutic target in poorly cohesive gastric carcinoma. J Transl Med. 2021;19(1):401. doi: 10.1186/s12967-021-03079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180(6):1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao Z, Su J. Progresses on three pattern recognition receptor families (TLRs, RLRs and NLRs) in teleost. Dev Comp Immunol. 2021;122:104131. doi: 10.1016/j.dci.2021.104131. [DOI] [PubMed] [Google Scholar]
- 75.Ho M-F, Weinshilboum RM. Immune mediator pharmacogenomics: TCL1A SNPs and estrogen-dependent regulation of inflammation. J Nat Sci. 2017;3(8):e416. [PMC free article] [PubMed] [Google Scholar]
- 76.Cirillo N, Prime SS. A scoping review of the role of metalloproteinases in the pathogenesis of autoimmune pemphigus and pemphigoid. Biomolecules. 2021;11(10):1506. doi: 10.3390/biom11101506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tulah AS, Beghé B, Barton SJ, Holloway JW, Sayers I. Leukotriene B4 receptor locus gene characterisation and association studies in asthma. BMC Med Genet. 2012;13:110. doi: 10.1186/1471-2350-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwon S-Y, Kim J-H. Role of leukotriene B4 receptor-2 in mast cells in allergic airway inflammation. Int J Mol Sci. 2019;20(12):E2897. doi: 10.3390/ijms20122897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim ND, Chou RC, Seung E, Tager AM, Luster AD. A unique requirement for the leukotriene B4 receptor BLT1 for neutrophil recruitment in inflammatory arthritis. J Exp Med. 2006;203(4):829–835. doi: 10.1084/jem.20052349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hartiala J, Li D, Conti DV, et al. Genetic contribution of the leukotriene pathway to coronary artery disease. Hum Genet. 2011;129(6):617–627. doi: 10.1007/s00439-011-0963-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nancey S, Boschetti G, Hacini F, et al. Blockade of LTB(4)/BLT(1) pathway improves CD8(+) T-cell-mediated colitis. Inflamm Bowel Dis. 2011;17(1):279–288. doi: 10.1002/ibd.21404. [DOI] [PubMed] [Google Scholar]
- 82.Hicks A, Monkarsh SP, Hoffman AF, Goodnow R. Leukotriene B4 receptor antagonists as therapeutics for inflammatory disease: preclinical and clinical developments. Expert Opin Investig Drugs. 2007;16(12):1909–1920. doi: 10.1517/13543784.16.12.1909. [DOI] [PubMed] [Google Scholar]
- 83.Gruzieva O, Xu C-J, Yousefi P, et al. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ Health Perspect. 2019;127(5):57012. doi: 10.1289/EHP4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bodor Z, Bodor K, Keresztesi Á, Szép R. Major air pollutants seasonal variation analysis and long-range transport of PM10 in an urban environment with specific climate condition in Transylvania (Romania) Environ Sci Pollut Res. 2020;27(30):38181–38199. doi: 10.1007/s11356-020-09838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuehner JN, Bruggeman EC, Wen Z, Yao B. Epigenetic regulations in neuropsychiatric disorders. Front Genet. 2019;10:268. doi: 10.3389/fgene.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turan S, Bastepe M. GNAS spectrum of disorders. Curr Osteoporos Rep. 2015;13(3):146–158. doi: 10.1007/s11914-015-0268-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammerschlag AR, Stringer S, de Leeuw CA, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49(11):1584–1592. doi: 10.1038/ng.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez-Nahm S, Mendez MA, Benjamin-Neelon SE, et al. DNA methylation of imprinted genes at birth is associated with child weight status at birth, 1 year, and 3 years. Clin Epigenetics. 2018;10:90. doi: 10.1186/s13148-018-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oh ES, Petronis A. Origins of human disease: the chrono-epigenetic perspective. Nat Rev Genet. 2021;22(8):533–546. doi: 10.1038/s41576-021-00348-6. [DOI] [PubMed] [Google Scholar]
- 90.Oh G, Koncevičius K, Ebrahimi S, et al. Circadian oscillations of cytosine modification in humans contribute to epigenetic variability, aging, and complex disease. Genome Biol. 2019;20(1):2. doi: 10.1186/s13059-018-1608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lussier AA, Zhu Y, Smith BJ, et al. Updates to data versions and analytic methods influence the reproducibility of results from epigenome-wide association studies. Epigenetics. 2022;66:1–16. doi: 10.1080/15592294.2022.2028072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1: “Summary of the studies in the meta-analysis for the association between Season of Birth and DNA methylation at birth and in children (age: 1 to 11 years)”. Baseline summary of participants from each of the individual cohorts.
Additional file 2. “Cohort-specific methods and declarations (cohorts listed in alphabetical order)”. Method description, Funding and Acknowledgements from the participating cohorts.
Additional file 3. “PACE analysis plan for Season of Birth and methylation profiles in children (6 June 2018)”. Analysis plan that was circulated amongst the participant cohorts for the Season of Birth study.
Additional file 4. Table S2: “Models used in this meta-analysis study”. Inflation (lambda) and bias information of the models. Table S3: “Comparison of magnitude and direction of the FDR-significant DNA methylation signals identified in the at-birth and childhood meta-analyses”.
Additional file 5. Table S4: “Direction of differential methylation of CpGs in DMRs of the at-birth and childhood analyses (preliminary analysis”).
Additional file 6. Table S5: “Genes mapped to DMP/DMR identified in at-birth samples of babies born in the latitude ≥ 50°N and some examples of their associations with biological functions”. Provides examples of known functional associations of genes mapped to significant CpG sites or differentially methylated regions identified in this study.
Additional file 7. Table S6: “Season of birth-associated differentially methylated regions (DMRs) at birth in babies born in latitudes < 50°N”. Differentially methylated regions and mapped genes identified in the at-birth samples of babies born in latitudes < 50°N (lower latitude subgroup analysis). Table S7: “Season of birth associated with differentially methylated regions (DMRs) in children born in latitudes ≥ 50°N”.
Additional file 8. Table S8: “Genes mapped to DMR identified in the childhood samples of children born in the latitude ≥ 50°N and some examples of their associations with biological functions”. Provides examples of known functional associations of genes mapped to significant differentially methylated regions identified in this study.
Additional file 9. “Trait Enrichment Analysis (EWAS Atlas)”. Trait names and the odds ratios for their association with CpG sites for all the models meta-analysed in this study.
Data Availability Statement
Individual cohort-level data can be obtained from the respective cohort (see Additional file 1: Table S1 and Additional file 2 for cohort details).