FIG. 1.
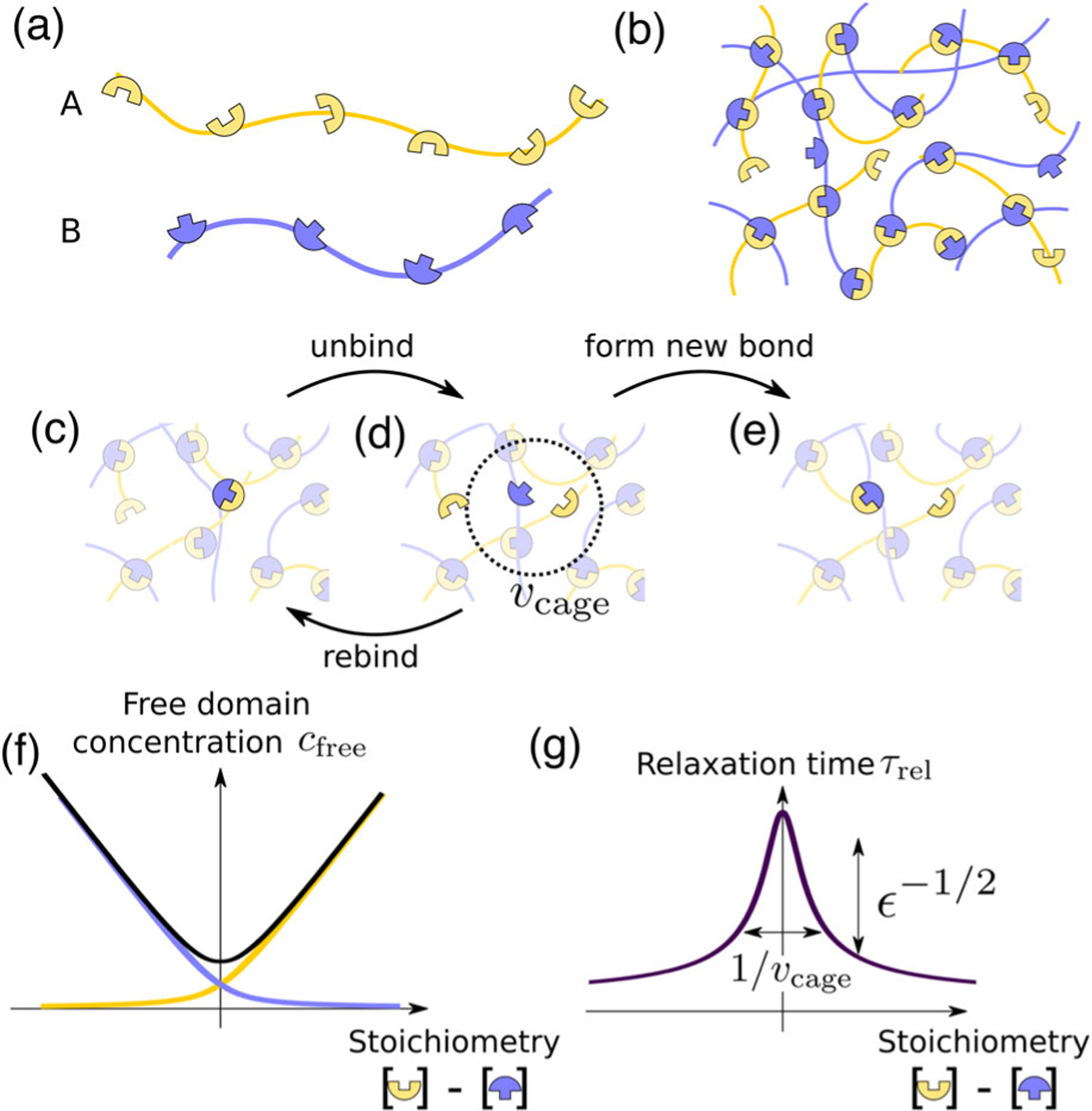
Stoichiometry controls the bond relaxation time of multivalent associative proteins. (a) Sketch of associative multivalent proteins, with complementary domains separated by flexible linkers. (b) Strong yet reversible binding between proteins leads them to condense into a network with most possible bonds formed. (c)–(e) Schematic of the bond relaxation mechanism. When two initially bound domains (c) unbind, the two are caged in a small volume (d). Two events can then occur: the initially bound domains can rebind, or, if a free domain is within reach, a new bond may form (e), which is the system’s basic relaxation mechanism. (f) Concentration of unbound domains of both types as a function of stoichiometry difference. (g) Relaxation time [Eq. (1)] corresponding to the process of unbinding and then rebinding with a new partner (c)–(e), as a function of stoichiometry difference. Here, .
