FIG. 2.
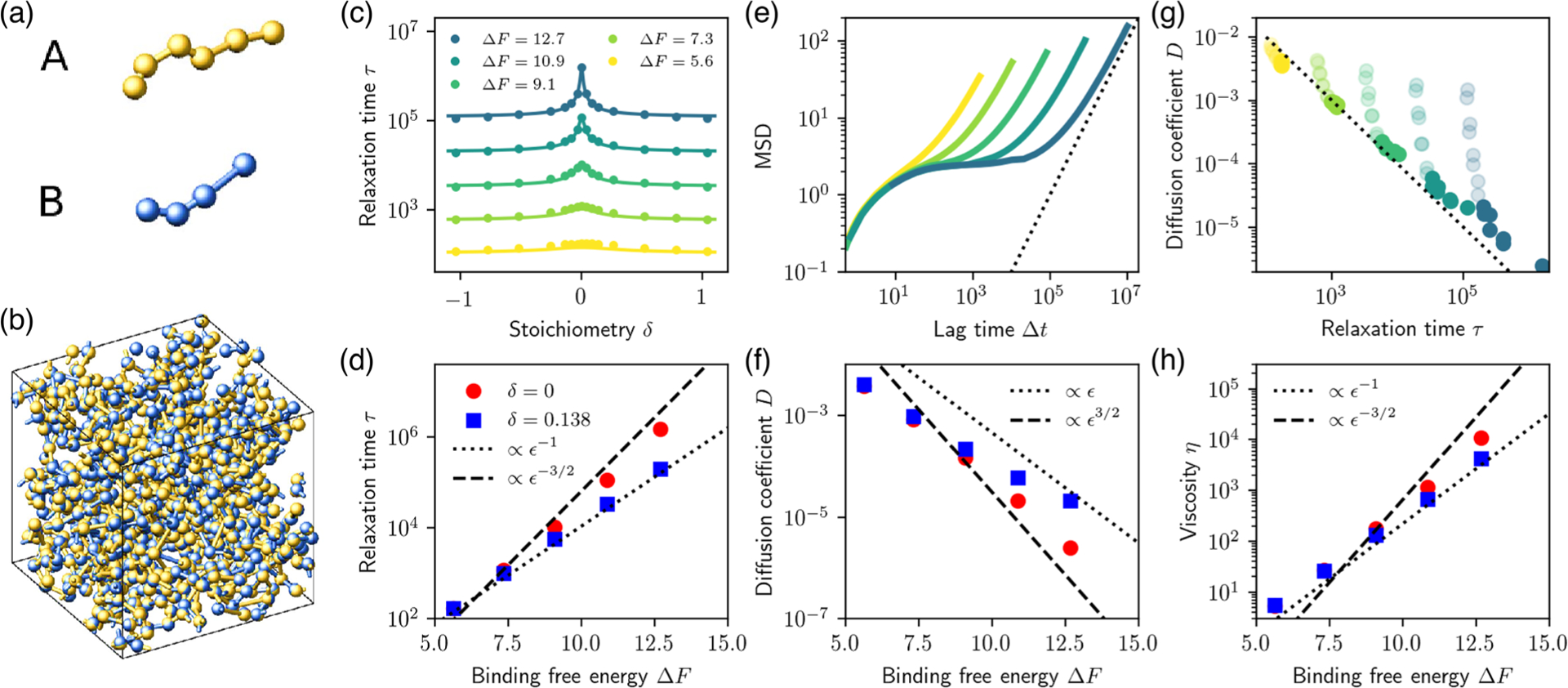
MD simulations reveal the importance of stoichiometry to the dynamical properties of the condensate. (a) MD model for the multivalent associative proteins. Colored spheres represent and domains. (b) Representative snapshot of the dense, network-forming liquid condensate. (c) Bond relaxation time (see text) as a function of stoichiometry for different binding strengths. Symbols indicate MD simulations; solid curves indicate theory [Eq. (1)] with estimated assuming full binding of the minority domains and with fitted values , and . (d) Bond relaxation time as a function of binding strength is consistent with predicted scaling for both equal and unequal stoichiometries [Eq. (1), Fig. 1(g)]. (e) Mean squared displacement (MSD) of individual domains as a function of time reveals diffusive scaling (dashed line) at long times (here ). (f) Diffusion coefficient of the minority species as a function of binding strength at equal and unequal stoichiometry. (g) Longtime diffusion coefficient plotted against bond relaxation time, for all values of and . The dotted black line indicates . Transparent circles correspond to systems where one component is in large excess, . (h) Viscosity, obtained using the Green-Kubo relation, as a function of binding strength, reflects the scaling of the bond relaxation time (d).
