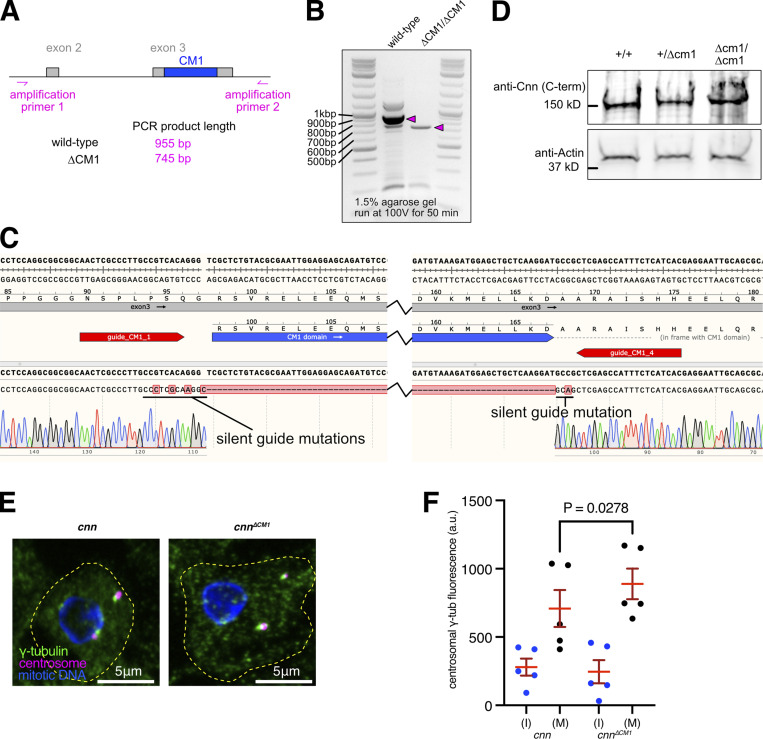
Figure S1.
Centrosomes from cnn∆CM1 mutants accumulate slightly more γ-tubulin than centrosomes from cnn null mutants. (A) Cartoon representation of region amplified to check for CM1 deletion mutants. (B) Gel showing DNA bands from PCR reactions when using the primers shown in A on either wild-type flies or CM1 deletion flies, as indicated. (C) Excerpt from SnapGene showing the sequencing result when using amplification primer 2 to sequence the PCR product generated using amplification primer 1 and amplification primer 2 on CM1 deletion flies. The position of the guide RNAs used when making the deletion is indicated. Note that silent mutations were introduced to prevent Cas9 from recutting the DNA after the recombination event. (D) Western blot of larval brain extracts from different genotypes, as indicated, probed with C-terminal anti-Cnn polyclonal antibodies. Note that the ∼8 kD difference in size between wild-type Cnn, which runs at ∼150 kD, and Cnn∆CM1 is not discernible. (E) Fluorescence images of mitotic Drosophila brain cells from either cnn or cnn∆CM1 mutant third instar larvae immunostained for γ-tubulin (green), mitotic DNA (blue), and Asl (centrioles, magenta). Note how the γ-tubulin signal in cnn∆CM1 mutant cells is also offset from the Asl signal, indicating that removing the CM1 domain affects the Cnn’s ability to form a proper centrosomal scaffold. Scale bars are 5 μm. (F) Graph showing average fluorescence intensities of interphase (blue dots) and mitotic (black dots) centrosomes from either cnn or cnn∆CM1 mutant brains (as indicated below). Each datapoint represents the average centrosome value from one brain. N = 5 for each dataset. Mean and SEM are indicated. Brains from the different genotypes were paired on slides (one slide per pair) allowing a two-sided paired t test to compare the mean values between mitotic centrosomes. Note that γ-tubulin accumulation at mitotic centrosomes is only slightly higher in cnn∆CM1 mutant cells, indicating that either the Cnn-dependent pool of Spd-2 is not an efficient recruiter of γ-TuRCs or that recruitment of the Cnn-dependent pool of Spd-2 is perturbed in cnn∆CM1 mutant cells or both. Source data are available for this figure: SourceData FS1.