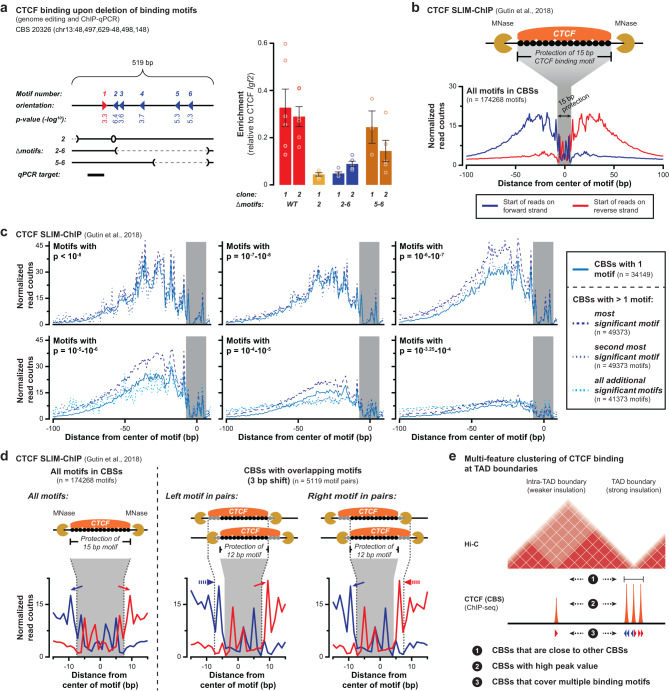
Fig. 2. Multiple binding motifs within CTCF binding sites contribute to CTCF binding.
a Removal of CTCF binding motifs within a CBS reveals that multiple motifs contribute to CTCF binding. Left: position of the 6 motifs within CBS 20326, with arrowheads indicating orientation and values indicating significance score. Below, motif deletions and the qPCR target are shown. Right: relative enrichment of CTCF binding as determined by ChIP-qPCR. Data are presented as mean values +/− SEM. Rings indicate individual measurements (n = at least 2 measurements each from 2 biological replicates). b Outline of the SLIM-ChIP assay to determine nuclease protection at CTCF motifs in WT mESCs. Top: SLIM-ChIP read-out, with black circles indicating protection at the CTCF binding motif. Bottom: normalized pile-up of SLIM-ChIP read start positions around all significant binding motifs in CBSs (200 bp window). The gray area overlaps the 15 bp motif where signal is reduced due to protection by CTCF binding. c Normalized pile-up of SLIM-ChIP signal (forward strand only, Supplementary Fig. 3a) for motifs sorted on their significance (different graphs) and their presence within different groups of CBSs (color coding in legend). Categories with fewer than 100 motifs have been excluded. P-values refer to motif significance score as determined using the MEME-suite (see Methods section). d Normalized pile-up of SLIM-ChIP signal for motif pairs that overlap with a 3 bp shift (encompassing ~6% of all motifs). Left: zoomed-in pile-up for all CTCF binding motifs. Middle and right: zoomed-in pile-up of the left and right motifs in overlapping pairs. Solid arrows highlight the two peaks with strong signal that surround the 15 bp binding motif. Dashed arrows highlight the 3 bp shift of the strong peaks, creating a 12 bp protected motif. Above, the impact of alternating CTCF binding on the left or right motif is indicated, which explains the presence of non-protected bases in both motifs, thereby confirming that they can both bind CTCF. e Summary of multi-feature enrichment of CTCF binding at strongly insulated TAD boundaries, as compared to sites elsewhere in the genome.