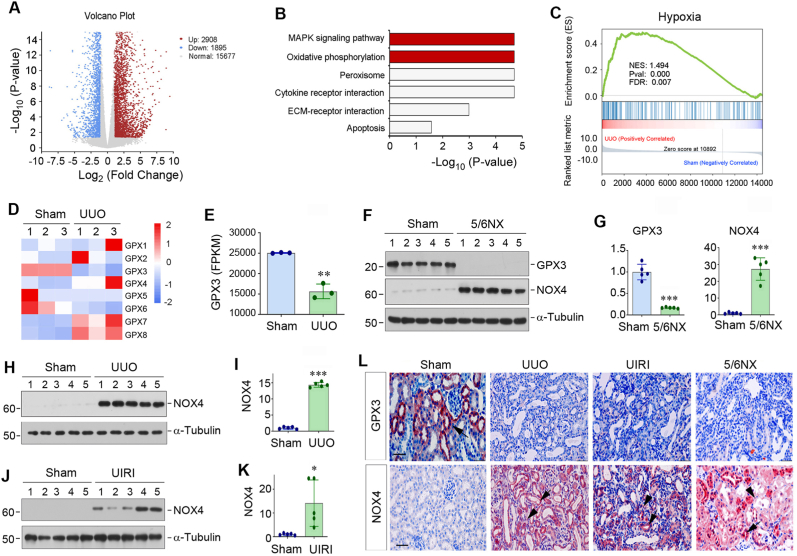
Fig. 1.
Gene expression profiling by RNA sequencing identifies loss of GPX3 as a pathologic feature of chronic kidney disease (CKD). (A) The volcano diagram of RNA-seq data displays the differentially expressed genes of sham and UUO kidneys. The red dots represent the genes up-regulated in UUO group, while blue dots show genes down-regulated. The grey dots denote mRNAs with no change in abundance between the two groups. (B) KEGG enrichment analysis reveals that several signaling pathways as indicated were enriched. (C) GSEA enrichment analysis shows that hypoxia was enriched in UUO group. NES, normalized enrichment score; FDR, false discovery rate. (D) The heat map shows differential expression of GPX family (GPX1-GPX8) genes in sham and UUO kidneys. (E) RNA-seq analysis shows the expression level (FPKM) of GPX3 in normal and UUO kidneys. FPKM, fragments per kilobase per million mapped reads. (F, G) Western blot analyses show the levels of GPX3 and NOX4 in different groups as indicated. Representative Western blot (F) and quantitative data (G) are shown. (H, I) Western blot analyses show the levels of NOX4 in sham and UUO mice. Representative Western blot (H) and quantitative data (I) are shown. (J, K) Changes of NOX4 expression in the kidney after UIRI. Representative Western blot (J) and quantitative data (K) are shown. *P < 0.05, **P < 0.01 and ***P < 0.001 versus sham (n = 5). (L) Immunohistochemical staining shows renal expression and localization of GPX3 (upper panel) and NOX4 (lower panel) proteins in different groups as indicated. Arrows indicate positive staining in renal tubular epithelial cells, whereas arrowheads indicate positive staining in renal interstitium. Scale bar, 50 μm.