Abstract
Human liver tissue is composed of heterogeneous mixtures of different cell types and their cellular stoichiometry can provide information on hepatic physiology and disease progression. Deconvolution algorithms for the identification of cell types and their proportions have recently been developed for transcriptomic data. However, no method for the deconvolution of bulk proteomics data has been presented to date. Here, we show that proteomes, which usually contain less data than transcriptomes, can provide useful information for cell type deconvolution using different algorithms. We demonstrate that proteomes from defined mixtures of cell lines, isolated primary liver cells, and human liver biopsies can be deconvoluted with high accuracy. In contrast to transcriptome-based deconvolution, liver tissue proteomes also provided information about extracellular compartments. Using deconvolution of proteomics data from liver biopsies of 56 patients undergoing Roux-en-Y gastric bypass surgery we show that proportions of immune and stellate cells correlate with inflammatory markers and altered composition of extracellular matrix proteins characteristic of early-stage fibrosis. Our results thus demonstrate that proteome deconvolution can be used as a molecular microscope for investigations of the composition of cell types, extracellular compartments, and for exploring cell-type specific pathological events. We anticipate that these findings will allow the refinement of retrospective analyses of the growing number of proteome datasets from various liver disease states and pave the way for AI-supported clinical and preclinical diagnostics.
Keywords: Fibrosis, Proteomics, Biomarker, Liver, Deconvolution, Computational Methods
Graphical Abstract
1. Introduction
Tissue samples and tissue-derived biomarkers are routinely used in clinical medicine and biological research. Tissues are comprised of heterogeneous mixtures of different cell types. More than 200 distinct cell types have been identified in humans and additional subtypes are frequently discovered as technology is improved [1], [2]. Transcript, protein, and metabolite signatures differ between cell types and, as such, variations in the proportions of different cell types and extracellular matrix (ECM) components impact the transcriptomic, metabolomic, and proteomic patterns of a given tissue sample. For instance, inflammation often results in an increased proportion of leukocytes, while ECM proteins proportionally increase in fibrosis.
Methodologies for determining composition of cell types have been developed for transcriptomic data using various deconvolution approaches. There are two categories of deconvolution approaches: partial deconvolution and complete deconvolution [3], [4], [5]. In partial deconvolution, prior information about the proportions of the different cell types or the signatures of isolated cell types are used to deconvolute the cell type composition. Complete deconvolution is made without such prior knowledge. Both partial and complete deconvolution have been used to infer cell type proportions and signatures from transcriptomics data. For instance, deconvolution of single cell transcriptomes has been used to study the infiltration of lymphocytes and other immune cells in the tumour microenvironment [6], [7]. Similarly, accounting for sample heterogeneity using deconvolution resulted in more accurate predictions of breast cancer recurrence and was able to guide clinical decision making [8]. However, while these strategies have been successfully applied to transcriptomic data, deconvolution of proteomic data has to our knowledge not yet been presented, at least in part because access to sufficiently large proteomic datasets has been limited. Furthermore, the number of proteins covered by state-of-the-art proteomics is significantly lower than the number of transcripts obtained from RNA sequencing methods and single cell-based proteomic methods lack the necessary depth and resolution for partial deconvolution methodologies [9], [10].
To overcome these knowledge gaps, we set out to investigate whether global proteomics data could be used for human cell type deconvolution. We compared a simple linear least squares method, as well as partial and complete deconvolution methods developed for transcriptomes [3], [11]. To establish the methodology, we first generated 20 blinded mixtures of cell lines with defined proportions, conducted bulk proteomics and used the resulting data to identify cell type composition. Although 90% of all proteins were found in all the cell lines, deconvolution accurately identified the proportions with correlation coefficients of 0.71–0.99. To our surprise, similar accuracies were obtained when deconvoluting proteomic data of defined mixtures of primary human liver cell types (hepatocytes, endothelial cells, Kupffer cells and stellate cells) obtained from proteomic repositories. Encouraged by this finding, we analysed the proteomes of human liver tissue samples from 56 donors undergoing Roux-en-Y gastric bypass surgery [12], [13] using complete deconvolution. Importantly, we found that an increased proportion of immune cells and stellate cells correlated with clinical immune response markers and altered ECM profiles, respectively. Combined, our results show that proteome deconvolution provides a molecular microscope for analyses of the composition of cell types and extracellular compartments, which can provide additional cell type-specific information using input data from new proteome collections and retrospective analyses of data deposited in proteomics databases [14].
2. Materials and methods
2.1. Proteomic analysis of HEK 293, Caco-2 and A549 cell lines
Samples, containing approximately 6 million cells of one cell line (HEK 293, Caco-2 or A549), or mixtures thereof with known proportions were prepared. The sample contents were blinded for the operator, and not decoded until deconvolution of the individual cell types in the mixtures was complete. The samples were prepared for proteomics analysis using a slightly modified MED-FASP protocol as previously described [15], [16]. In short, the cell samples were chemically lysed, protein concentration was measured using the tryptophan fluorescence method [17] and proteolytic digestion was subsequently performed with Lys C and trypsin. The peptides were injected on an UltiMate 3000 RSLC nano system coupled to a QExactive HF mass spectrometer (Thermo-Fisher Scientific, Palo Alto, CA, USA) using a Top15 method (full MS followed by 15 ddMS2 scans). The data were analysed using MaxQuant version 1.6.0.16 with the complete human proteome extracted from UniProtKB version 2015 June. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [18] with the dataset identifier PXD027282.
Extended method descriptions are available in the Supporting Information.
2.2. Pre-processing of proteome data for deconvolution
Protein concentrations were calculated using the Total Protein Approach [16]. Total protein values were used as input for the deconvolution algorithms. The COCKTAIL dataset from human liver samples contains samples analysed on different occasions and batch effects were controlled for using geometrical mean centring as described previously [13].
2.3. In silico mixing of primary human liver cells
Endothelial cells (EC), hepatocytes (HC), Kupffer cells (KC) and stellate cells (SC) make up the majority of cells in the liver. Using cell type-specific data of these cell types from human liver [15], an in silico mixture of 100 samples was created. The proportions were assigned using a random uniform distribution divided by the sum of the proportions, so the sum was equal to one. The concentrations of the proteins in each sample were calculated by multiplying the proportions of the cell type with the protein-specific variation obtained from a normal distribution using cell type-specific means and standard deviations. The calculated concentrations of the 100 in silico mixed samples were then used by the algorithms to determine the cell type proportions.
2.4. Computational deconvolution methodologies
To investigate whether algorithms previously developed to deconvolute transcriptomics data could be applied to proteome datasets, we tested MuSiC [11], a partial deconvolution algorithm that uses cell type-specific information to quantify cell type compositions from bulk data, as well as Linseed [3], which performs complete deconvolution and does not require input of cell type-specific data, but instead requires annotation of the cell types to be identified. For the latter, we used single cell RNA sequencing data from human liver and the UniProt database [19], [20] to annotate the identified cell types for tissue when using Linseed. A linear methodology using a constrained linear least squares solver was developed in Matlab to investigate how the MuSiC and Linseed algorithms compared to a simple linear methodology without filtering, scaling, or identification of marker proteins. Extended method descriptions are available in the Supporting Information.
2.5. Proteome data set of human liver tissue
The human liver proteomes were obtained from the COCKTAIL study with trial registration number NCT02386917 [12], [13]. The 56 patients underwent Roux-en-Y gastric bypass surgery during which liver biopsies were collected for omics investigation. Anthropometric measurements and blood samples for clinical chemistry analyses were also collected. The study protocol was approved by the Regional Committee for Medical and Health Research Ethics (2013/2379/REK sørøst A), and all patients signed a written informed consent.
2.6. Proteome data set of isolated human liver cell types
The proteomes of isolated hepatocytes and non-parenchymal cells (NPCs) from human liver [15] were extracted from the PRIDE repository [21] using the identifier PXD012615.
3. Results
3.1. Deconvolution of mixtures of cell lines
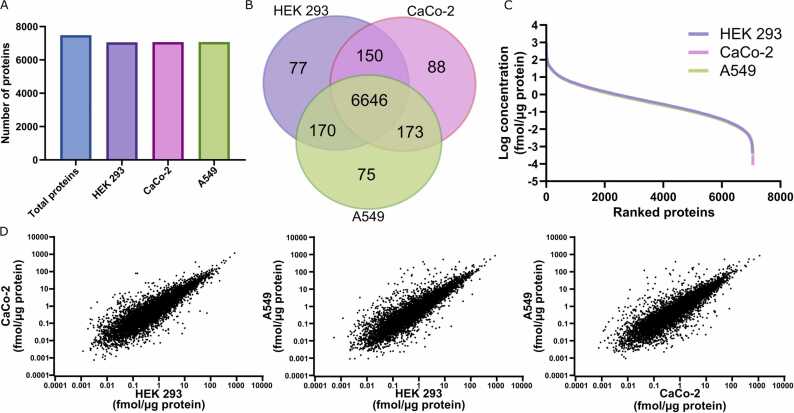
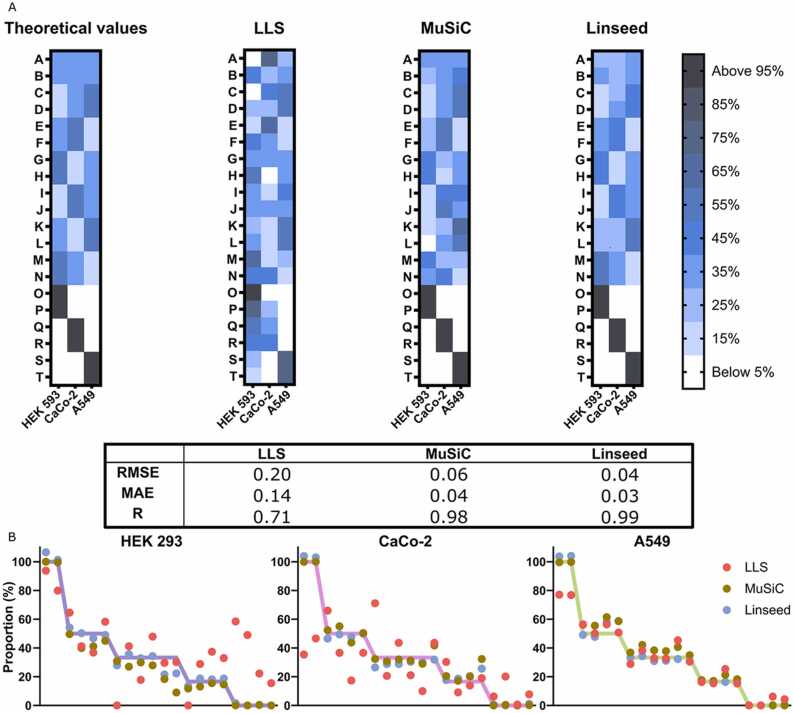
To establish the deconvolution methodology on proteomic data, we first used three cell lines of different tissue origins (HEK 293: human embryonic kidney, Caco-2: human colorectal adenocarcinoma, and A549: human adenocarcinomic alveolar basal epithelium). A total of 20 samples, consisting of six samples containing only one of the three selected cell lines (in duplicates), plus 14 mixtures of the three cell lines with different stoichiometry were generated and subjected to global proteomics analysis. In total, 7379 proteins were identified. More than 90% of those were detected in all three cell lines (Fig. 1A, B). We furthermore confirmed that the distribution of the protein concentrations was comparable between the cell lines (Fig. 1C-D). The mixtures of the cell lines in different proportions were used with the simple constrained LLS algorithm, the partial deconvolution algorithm MuSiC, and the complete deconvolution algorithm Linseed, to quantify cell stoichiometries. In the case of MuSiC (partial deconvolution), we used the six single cell line samples to produce the specific proteomic signatures of each of the cell lines. The LLS method could not properly decipher the different cell proportions in any of the samples (R=0.71). However, despite the substantial overlap of proteomic signatures between cell lines, both partial deconvolution using MuSiC (R=0.98) and complete deconvolution using Linseed (R=0.99), accurately predicted the cell line stoichiometry across samples (Fig. 2A). No systematic under- or over-prediction was observed (Fig. 2B).
Fig. 1.
Global proteomes of cell lines from different tissues. The proteomes of three cell lines of different tissue origin were analysed. A, In total, 7043 (HEK 293), 7057 (Caco-2) and 7064 (A549) proteins were identified. B, Most proteins (96.7%) were shared between the cell lines. C, The overall ranking of protein concentrations was highly similar between cell lines. D, Correlation plots of protein expression levels were also similar, but distinct differences were evident.
Fig. 2.
Proteome deconvolution of cell lines results in accurate deconvolution of cell stoichiometries. The proteomes of individual and predetermined mixtures of cell lines were determined. A total of 20 (A-T) samples were used where six (O-T) contained only one of each cell line and the remaining samples (A-N) were mixtures; all samples were analysed blinded and in duplicate. A, Heat maps showing the prediction accuracy of the cell type proportions generated by three different deconvolution approaches. Root mean square error (RMSE), mean absolute error (MAE) and Pearson’s correlation (R) for the three algorithms are shown. B, Proportions of the three different cell lines in the 20 samples. Dots indicate predicted values by the different algorithms while lines show the true values.
3.2. Deconvolution of mixtures of primary human liver cells
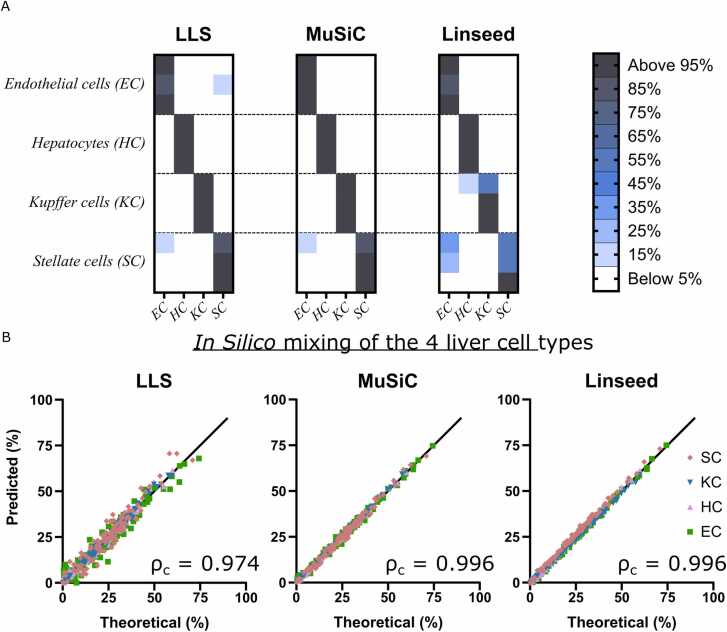
Since the algorithms properly estimated cell proportions using the proteomics data from cell lines, we next investigated whether the same approach could be applied to primary human liver cells, that is cells from the same tissue. Samples isolated from three donors [15], representing each of the four major liver cell types (hepatocytes, stellate cells, liver sinusoidal endothelial cells, and Kupffer cells), were used for proteome-informed deconvolution. All three algorithms correctly found that each of the samples contained mostly one cell type (Fig. 3A). We continued by producing 100 in silico mixtures containing the four major cell types in different proportions. All three algorithms accurately deconvoluted the mixtures; however, MuSiC and Linseed (both ρc=0.996) produced slightly more accurate estimates of cell proportions than LLS (ρc=0.974; Fig. 3B), corroborating the results obtained in cell lines.
Fig. 3.
Proteome deconvolution of the four major liver cell types in human liver. The proteomes of liver sinusoidal endothelial cells (EC), hepatocytes (HC), Kupffer cells (KC) and stellate cells (SC) were determined previously [15]. A, Heat maps showing that the different liver cell types can be accurately deconvoluted based on proteomic data. B, Predicted versus true cell type proportions in 100 mixtures of the four cell types. Lin's concordance correlation coefficient (ρc) is indicated for each of the three algorithms.
3.3. Deconvolution of human liver tissue
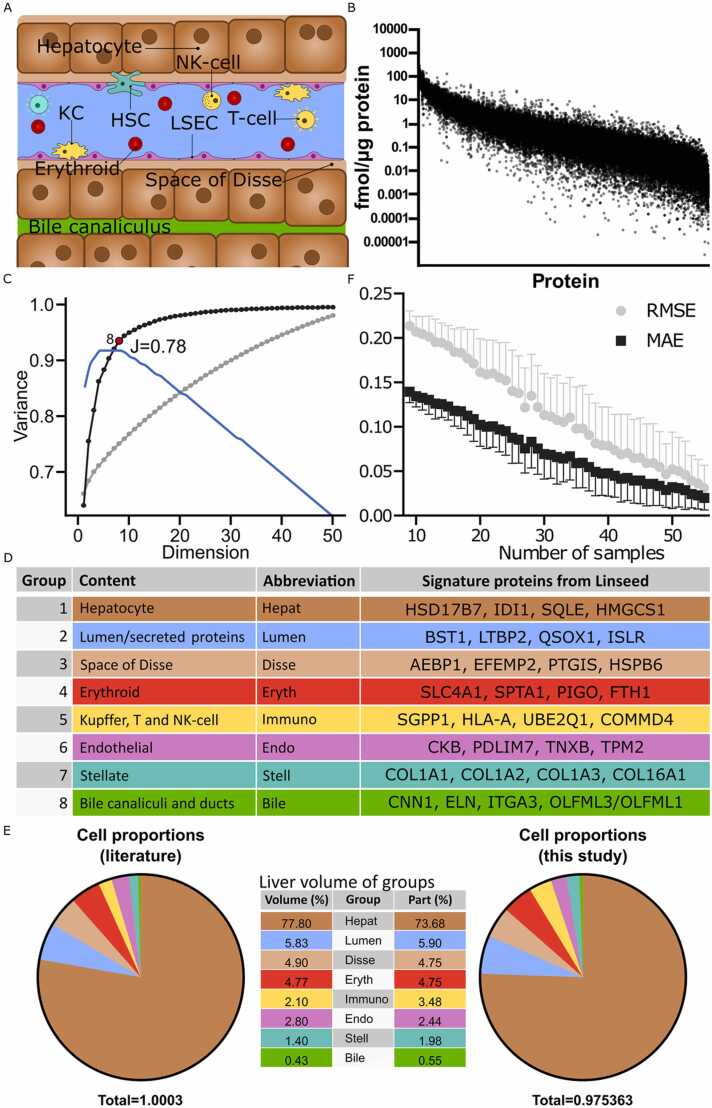
Based on these results, we proceeded to deconvolute proteomes obtained from liver tissue of 56 individuals undergoing Roux-en-Y gastric bypass surgery in the COCKTAIL study [12], [13]. Since liver tissue is comprised by more cell types (e.g., blood/immune cells) than the four archetypal liver cells investigated in Fig. 3, we used the complete deconvolution algorithm Linseed, as partial deconvolution requires prior knowledge about the purified cell type proteomes to estimate the proportions in bulk tissue. Our aim was therefore to deconvolute the cell stoichiometry of the four archetypal liver cell types, as well as various populations of liver resident and circulating immune cells (Fig. 4A). Overall, the quantified proteins varied by 2–3 orders of magnitude between individuals (Fig. 4B). We used singular value decomposition (SVD) to estimate the number of cell types by inferring the number of linearly independent components [3]. Notably, 94% of the variance could be explained by the first eight components with a Youden index of 0.78 (Fig. 4C). Annotation of these components using data from single cell transcriptomics and the UniProt database [19], [20] revealed that component one, which alone explained 63% of variance, corresponded to hepatocytes. Signature proteins for this group were identified as HSD17B7, IDI1, SQLE and HMGCS1, which encode hepatocytic enzymes with important roles in steroid and cholesterol metabolism (Fig, 4D). Notably, typical hepatocyte markers, such as major drug metabolizing enzyme CYP3A4, were not part of the signature proteins. Signature proteins for deconvolution should increase linearly with the number of cells. However, CYP3A4 expression is heavily influenced by both genetic and environmental factors [22] and, thus does not constitute a suitable signature protein for deconvolution.
Fig. 4.
Proteome deconvolution can accurately reveal the composition of cell types of human liver biopsies. The proteomes from 56 human liver tissue samples were analysed using complete deconvolution. A, Schematic drawing showing cell and extracellular compartments identified by deconvolution. Colour codes are harmonized between fig A, D and E. B, Variability in liver protein expression. C, Variance explained cumulatively for each Singular Value Decomposition component for unfiltered (all proteins; grey) and filtered (collinear proteins; black). The first eight components explain 94% of all variance for the filtered data and had a Youden index of 0.78. D, The eight compartments identified in C and examples of their signature proteins. E, Average estimated proportions (by volume) in human liver (n = 56) of the eight compartments were in excellent agreement with the literature values [23]. F, Variability as a function of sample size.
Components two (adding 12% to the explained variance) and three (adding 6%) corresponded to extracellular components in the lumen and space of Disse. Importantly, this variability will be missed when using conventional transcriptomic rather than proteomic deconvolution. Groups 4–7 corresponded to erythroid cells, immune cells, liver sinusoidal endothelial cells and stellate cells, respectively, while the last component was identified as a bile canalicular compartment. Comparison of the abundance of the annotated groups with previously published data on the relative composition of hepatic cell types [23] demonstrated an excellent alignment with the corresponding fractions of liver volume occupied by the respective compartments further corroborating the accuracy of the deconvolution method on heterogenous patient-derived samples (Fig. 4E). As expected, predictive accuracy increased with sample size (Fig. 4F). However, even when sample number were decreased from 56 to 20, the mean absolute error (MAE) and the root mean square error (RMSE) of predictions remained < 0.1 and < 0.2, respectively (Fig. 4E). This result demonstrates that complete deconvolution based on proteomes even constitutes a suitable and reliable approach also for smaller sample sets, thus increasing applicability to studies for which only around 20 samples are available.
3.4. Correlation between liver cell composition and clinical parameters
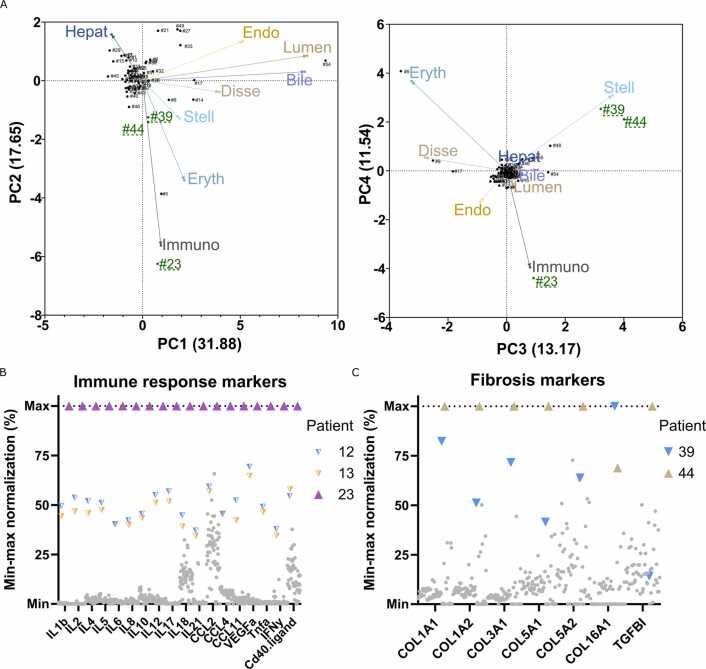
Next, we analysed inter-individual differences in liver cell composition. Most liver samples had comparable cell compositions and formed one large cluster in eigenvector-based multivariate analyses (Fig. 5A). However, ten of the 54 liver samples were identified as outliers. In sample #23, we identified elevated levels of immune cells. Comparison with blood samples collected at the time of the sampling of the liver biopsies revealed that patient #23 had the highest values of all patients for pro-inflammatory interleukins, chemokines, and TNF-α (Fig. 5B). These results suggest that the systemic inflammation identified in peripheral blood is closely aligned with an increase in hepatic immune cell composition. Interestingly, while elevated levels of immune cells revealed by the complete deconvolution agreed with clinical immune response markers, the patient had a hsCRP below 10 mg/l before surgery and no clinically manifest symptoms of inflammation, suggesting that deconvolution can provide additional information that goes beyond what is obtained by conventional clinical chemistry testing.
Fig. 5.
Patients with deviating cellular proportions have altered clinical chemistry. A, Principal component analysis biplots of the estimated cell proportions for the 56 human liver samples. Green indicates livers that deviate from the average liver cell type proportions. Arrows indicate the loading vectors. The numbers in parentheses represent the percentage of variability explained by each principal component. B, Immune response markers in patient blood samples (n = 56). Note the agreement between the enhanced immune cell proportion identified by deconvolution and the high levels of inflammatory markers in patient #23. C, Fibrosis markers identified in the patient proteomes (n = 56). Note the agreement between the increased stellate cell proportions and the altered composition of extracellular matrix proteins in patients’ number #39 and #44. Coloured symbols indicate outlier patients of interest, while grey symbols denote the other patients.
Similarly, patients #39 and #44 showed elevated levels of hepatic stellate cells, which play important roles in the initiation and progression of fibrosis [24]. None of these patients had FIB-4 fibrosis scores above the clinically relevant cut-off value (>2.67), indicating that no patient had advanced fibrosis [25], which was also confirmed during the surgery. However, the composition of ECM is altered progressively during fibrosis (i.e., already in early non-advanced stages) with an increase in fibrillar collagen (e.g., collagen types I and III) and a decrease in fibril associated collagens with interrupted triple helices (FACIT; e.g. collagen types XII and XIV). We therefore investigated ECM protein composition (Suppl Fig. 1A) and found that the ECM signatures for these patients indeed formed a separate cluster characterized by elevated levels of fibrillar collagens consistent with the alterations in stellate cell abundance (Fig. 4C). These results indicate that complete deconvolution of bulk proteomic data can flag patients with altered ECM protein composition at early stages that are missed using conventional FIB-4 scoring, thus providing a useful complement for diagnostics and in-depth investigations into disease phenotypes.
4. Discussion
Deconvolution algorithms have previously been used to obtain tissue-specific information about cell type composition from bulk transcriptomic data [3], [11], [26]. Here we show that also proteomic data, which typically contains less data points than transcriptomes, can be used to accurately deconvolute cell-specific proportions in complex mixtures. The results show that it is possible to deconvolute proteomic data from cell lines, mixtures of primary cells, and human liver tissue, providing new information on tissue composition and allowing the identification of cell type-specific disease biomarkers from complex mixtures. Furthermore, in contrast to transcriptome data, proteomic deconvolution also allows to obtain tissue proportions of important extracellular components, such as matrix proteins.
In the first set of experiments, the proteomes of blinded mixtures of three cell lines from different tissues were deconvoluted. The cell lines shared most proteins and only around 80 proteins were unique, representing 3% of the total proteome. This may seem like a small number, but previous findings suggest that less than 5% of all gene products are tissue specific [27]. Hence, deconvolution based on only cell type-specific markers could be challenging and too noisy. Despite the substantial overlap, both partial (MuSiC) and complete (Linseed) deconvolution resulted in highly accurate inference of cell type composition with root mean square errors (RMSE) similar to those previously observed in unrelated transcriptomic data [11]. These results indicate that despite the smaller data size of proteomic datasets, accuracy does not decrease. While bias for certain cell types has previously been observed in deconvolution studies using transcriptomic data, likely due to cell type-specific differences in RNA content [3], we did not observe any indication of bias in our proteomic deconvolution. Additionally, Lin's concordance correlation coefficient for both partial and complete deconvolution resulted in close to perfect correlations, indicating that both deconvolution methods were able to determine the cell type proportions in mixtures of different liver cell types with high accuracy.
For the identification of multiple cell types and their proportions in tissues, complete deconvolution algorithms are preferred as they do not require prior knowledge of the proteomes of each cell type. Another advantage is that tissue proteomes provide a snapshot of the different cell types in their natural environment compared to isolated cell types used in partial deconvolution. Therefore, we proceeded with complete deconvolution of data from the human liver tissue. An important aspect of complete deconvolution methods is that they require an estimate of the number of constituents, which can be cell types or distinct extracellular compartments, present in the sample [20], [27], [28]. To mitigate this problem, we used Singular Value Decomposition (SVD) to predict the number of constituents. Importantly, SVD showed that around 95% of the variance could be explained by eight proteome groups, which could be annotated to liver-specific cell types and compartments and corresponded to previously published proportions of liver cell types and extracellular tissue components [23]. These results provide an orthogonal validation of the approach and paved the way for analyses of inter-individual differences in liver proteome compartmentalization.
While liver compositions were similar among most of the 56 individuals analysed, some patients displayed elevated numbers of immune cells or stellate cells, characteristic of hepatic inflammation and fibrosis, respectively. Importantly, the patient with elevated levels of immune cells passed the standard clinical evaluation prior to surgery, had low level of hsCRP, and displayed no apparent signs of inflammation during surgery. Nevertheless, this patient had the highest levels of all 56 patients for 18 of the most relevant immune response markers. This relationship between elevated hepatic immune cell numbers and increased cytokine and chemokine levels are in alignment with a patient history of weekly cholestatic episodes [29]. Two other patients (patients #12 and #13), who showed significantly elevated levels of immune response markers did not exhibit increased immune cell numbers in the liver. These results are important as they indicate that liver proteome deconvolution can distinguish systemic and liver-specific inflammation, which can allow the refinement of further clinical interventions.
Two patients showed an increase in stellate cell numbers but were not classified as having clinically manifestation of hepatic fibrosis based on FIB-4 scoring, which considers demographic factors, thrombocytopenia as well as blood-based parameters of liver injury. A sub-analysis of all extracellular matrix proteins found in the liver at different stages of fibrosis [30] found that proteins associated with fibrosis, such as fibrillar collagen, were elevated in these two patients. Notably, while FIB-4 scoring has been shown to be superior to other non-invasive algorithms for the prediction of fibrosis in NAFLD [31] with performance similar to liver biopsies (AUC, 0.71–0.89), its accuracy for the progression of fibrosis is considerably lower (AUC, 0.57–0.81) [32]. Thus, observations of elevated stellate cell numbers correlate with clinical markers of altered ECM composition characteristic of fibrosis development and can provide complementary information that might refine risk stratification based on conventional non-invasive markers.
In conclusion, our results show that proteome deconvolution provides an effective tool for investigating the cell type composition human liver. A unique feature of proteome deconvolution is that it also provides valuable information about extracellular compartments. Importantly, complete deconvolution of patient liver biopsies allowed to identify individuals with altered tissue composition, which correlated with haematological parameters and established histological markers of liver fibrosis. Combined, this provides an opportunity for the analysis of disease mechanisms and the identification of diagnostic markers in disease-specific proteome collections.
Funding
This work was supported by the Swedish Research Council (grant numbers 2822 and 01951) to P.A.
CRediT authorship contribution statement
Conceptualization, N.H., D.Y. M.Ö. and P.A.; Methodology, N.H. and D.Y.; Software, N.H. and D.Y.; Validation, N.H., D.Y. and P.A.; Formal analysis, N.H., D.Y. M.Ö., C.W. and P.A.; Investigation, N.H., D.Y., M.Ö., C.W., C.K., R.J., J.H., A.Å, V.M.L. and P.A; Resources, P.A. D.Y., R.J., J.H. and A.Å.; Data curation, N.H.; Writing — original draft preparation, N.H. and P.A.; Writing — review and editing, N.H., D.Y., M.Ö., C.W., C.K., R.J., J.H., A.Å., V.M.L and P.A., Visualization, N.H., V.M.L and P.A.; Supervision, D.Y. and P.A.; Project administration, N.H. and P.A.; Funding acquisition, P.A. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
Volker M. Lauschke is CEO and shareholder of HepaPredict AB, as well as co-founder and shareholder of PersoMedix AB. Rasmus Jansson-Löfmark is an AstraZeneca employee and own shares in AstraZeneca, while Christine Wegler is a former AstraZeneca employee. The other authors have no competing interests to declare.
Acknowledgments
We would like to thank Dr. Andrea Treyer for supplying and mixing the cell lines, Professor Pierre Flener, Dr. Patrik Lundquist, Dr. Maria Karlgren, Dr. Varun Maturi, Sarah McComas, Signe Klinting, Evgeniya Mickols, and the members of the COCKTAIL study for helpful discussions, Professor Jacek Wisniewski and Katharina Zettl for producing the COCKTAIL proteomics data, Professor Göran Frenning for his mathematical support, and the participants, surgical staff and study personnel working on the COCKTAIL study at Vestfold Hospital Trust.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.08.037.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Giladi A., Amit I. Immunology, one cell at a time. Nat N. 2017;547:27. doi: 10.1038/547027a. [DOI] [PubMed] [Google Scholar]
- 2.Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., et al. The human cell atlas. eLife. 2017;6 doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaitsev K., Bambouskova M., Swain A., Artyomov M.N. Complete deconvolution of cellular mixtures based on linearity of transcriptional signatures. Nat Commun. 2019;10:2209. doi: 10.1038/s41467-019-09990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadel B.B., Oliva M., Shou B.L., Mitchell K., Ma F., Montoya D.J., Mouton A., Kim-Hellmuth S., Stranger B.E., Pellegrini M., et al. Systematic evaluation of transcriptomics-based deconvolution methods and references using thousands of clinical samples. Brief Bioinform. 2021;22:bbab265. doi: 10.1093/bib/bbab265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Im Y., Kim Y. A comprehensive overview of RNA deconvolution methods and their application. Mol Cells. 2023;46:99–105. doi: 10.14348/molcells.2023.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapuente-Santana Ó., Eduati F. Toward systems biomarkers of response to immune checkpoint blockers. Front Oncol. 2020:10. doi: 10.3389/fonc.2020.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma A., Merritt E., Hu X., Cruz A., Jiang C., Sarkodie H., Zhou Z., Malhotra J., Riedlinger G.M., De S. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. bioRxiv. 2019 doi: 10.1101/698845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elloumi F., Hu Z., Li Y., Parker J.S., Gulley M.L., Amos K.D., Troester M.A. Systematic bias in genomic classification due to contaminating non-neoplastic tissue in breast tumor samples. BMC Med Genom. 2011;4:54. doi: 10.1186/1755-8794-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manzoni C., Kia D.A., Vandrovcova J., Hardy J., Wood N.W., Lewis P.A., Ferrari R. Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences. Brief Bioinform. 2018;19:286–302. doi: 10.1093/bib/bbw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D., Eraslan B., Wieland T., Hallström B., Hopf T., Zolg D.P., Zecha J., Asplund A., Li L., Meng C., et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Syst Biol. 2019;15 doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019:10. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjelmesæth J., Åsberg A., Andersson S., Sandbu R., Robertsen I., Johnson L.K., Angeles P.C., Hertel J.K., Skovlund E., Heijer M., et al. Impact of body weight, low energy diet and gastric bypass on drug bioavailability, cardiovascular risk factors and metabolic biomarkers: protocol for an open, non-randomised, three-armed single centre study (COCKTAIL) BMJ Open. 2018:8. doi: 10.1136/bmjopen-2018-021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegler C., Garcia L.P., Klinting S., Robertsen I., Wiśniewski J.R., Hjelmesæth J., Åsberg A., Jansson‐Löfmark R., Andersson T.B., Artursson P. Proteomics-Informed Prediction of Rosuvastatin Plasma Profiles in Patients with a Wide Range of Body Weight. Clin Pharmacol Ther N/a. 2021 doi: 10.1002/cpt.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S., et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ölander M., Wiśniewski J.R., Artursson P. Cell-type-resolved proteomic analysis of the human liver. Liver Int. 2020;40:1770–1780. doi: 10.1111/liv.14452. [DOI] [PubMed] [Google Scholar]
- 16.Wiśniewski J.R., Rakus D. Multi-enzyme digestion FASP and the ’Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. J Proteom. 2014;109:322–331. doi: 10.1016/j.jprot.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Wiśniewski J.R., Gaugaz F.Z. Fast and sensitive total protein and peptide assays for proteomic analysis. Anal Chem. 2015;87:4110–4116. doi: 10.1021/ac504689z. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacParland S.A., Liu J.C., Ma X.-Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein K., Thomas M., Winter S., Nussler A.K., Niemi M., Schwab M., Zanger U.M. PPARA: A Novel Genetic Determinant of CYP3A4 In Vitro and In Vivo. Clin Pharmacol Ther. 2012;91:1044–1052. doi: 10.1038/clpt.2011.336. [DOI] [PubMed] [Google Scholar]
- 23.Bogdanos D.P., Gao B., Gershwin M.E. Liver Immunology. Compr Physiol. 2013;3:567–598. doi: 10.1002/cphy.c120011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C.-Y., Yuan W.-G., He P., Lei J.-H., Wang C.-X. Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol. 2016;22:10512–10522. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Younossi Z.M., Noureddin M., Bernstein D., Kwo P., Russo M., Shiffman M.L., Younes Z., Abdelmalek M. Role of noninvasive tests in clinical gastroenterology practices to identify patients with nonalcoholic steatohepatitis at high risk of adverse outcomes: expert panel recommendations. J Am Coll Gastroenterol ACG. 2021;116:254–262. doi: 10.14309/ajg.0000000000001054. [DOI] [PubMed] [Google Scholar]
- 26.Avila Cobos F., Alquicira-Hernandez J., Powell J.E., Mestdagh P., De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015:347. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 28.Aizarani N., Saviano A., Sagar, Mailly L., Durand S., Herman J.S., Pessaux P., Baumert T.F., Grün D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M., Cai S.-Y., Boyer J.L. Mechanisms of bile acid mediated inflammation in the liver. Mol Asp Med. 2017;56:45–53. doi: 10.1016/j.mam.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baiocchini A., Montaldo C., Conigliaro A., Grimaldi A., Correani V., Mura F., Ciccosanti F., Rotiroti N., Brenna A., Montalbano M., et al. Extracellular Matrix Molecular Remodeling in Human Liver Fibrosis Evolution. PLoS ONE. 2016:11. doi: 10.1371/journal.pone.0151736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A.G., Lydecker A., Murray K., Tetri B.N., Contos M.J., Sanyal A.J. Comparison of noninvasive markers of fibrosis in patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2009;7:1104–1112. doi: 10.1016/j.cgh.2009.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J., Vali Y., Boursier J., Spijker R., Anstee Q.M., Bossuyt P.M., Zafarmand M.H. Prognostic accuracy of FIB-4, NAFLD fibrosis score and APRI for NAFLD-related events: a systematic review. Liver Int. 2021;41:261–270. doi: 10.1111/liv.14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.