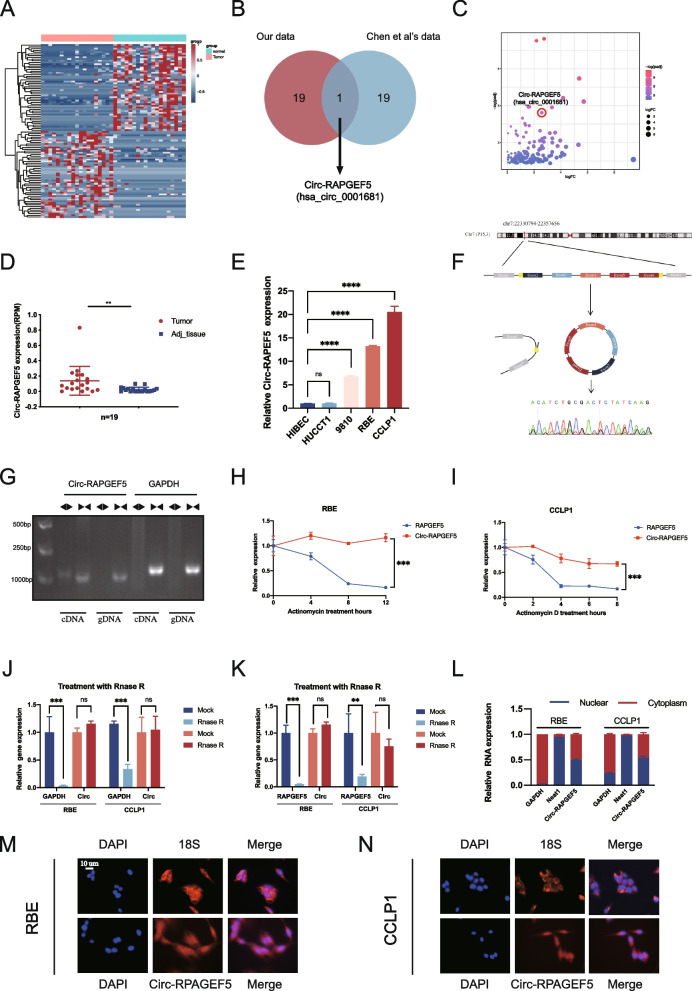
Fig. 1.
Circ-RAPGEF5 identification and validation in ICC cells and tissues. A Heatmap of the top 50 upregulated and downregulated expression circular RNAs between ICC tissues and adjacent normal tissues according to the bulk RNA sequencing of 19 patients. B Venn diagram illustrating the overlap of top 20 upregulated expression circular RNAs in our data and Chen et al.’ s data, only Circ-RAPGEF5 was identified. C Volcano plot of upregulated expressed circular RNAs from 19 paired samples. D Relative Circ-RPAGEF5 expression in 19 ICC tumor tissues and matched adjacent normal tissues. E qRT-PCR analysis of the relative expression levels of Circ-RAPGEF5 in a panel of four human ICC cell lines (HUCCT1, 9810, RBE, and CCLP1) and a normal biliary epithelial cell line, HIBEC. F The schematic illustration showed the back-splicing of Circ-RAPGEF5, and Sanger sequencing validated the splicing site. G Circular formation of Circ-RAPGEF5 was confirmed through PCR and agarose gel electrophoresis using divergent and convergent primers in both gDNA and cDNA samples of RBE. GAPDH was used as a negative control. H-I qRT-PCR analysis of Circ-RAPGEF5 and linear RAPGEF5 expression levels after Actinomycin D treatment in RBE and CCLP1. J-K qRT-PCR analysis of Circ-RAPGEF5 compared to linear GAPDH and RAPGEF5 expression levels after RNase R treatment in RBE and CCLP1. L-N Subcellular fractionation and FISH assays indicated that Circ-RAPGEF5 was localized in both the cytoplasm and nucleus of RBE and CCLP1. All data are presented as the means ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001