Figure 2.
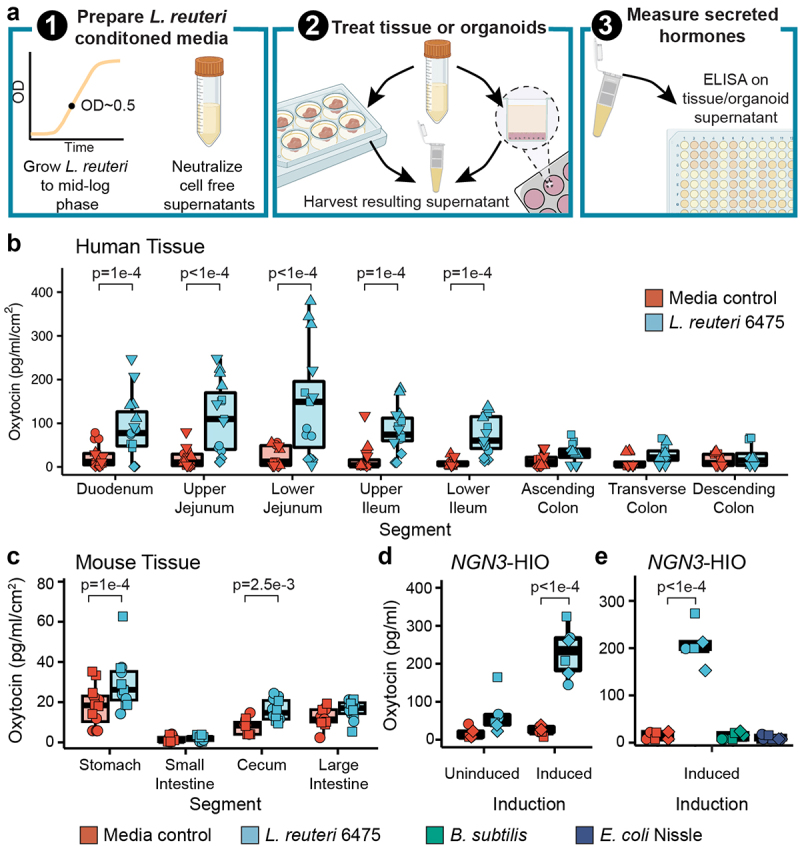
L. reuteri-conditioned medium promotes release of oxytocin from the gut epithelium. a) workflow of secretion assays. L. reuteri-conditioned medium is prepared by growing L. reuteri to mid-log phase, spinning down the bacterial cultures, and harvesting the supernatant. The supernatant is then neutralized to ~pH 7 and filter sterilized to remove cells while maintaining released products. For ex vivo tissue assays, L. reuteri-conditioned medium is placed onto intestinal tissue segments that have been washed free of luminal contents/fecal material. The tissue is then incubated with the L. reuteri-conditioned medium or L. reuteri growth medium control for 3 hours at 37°C with 5% CO2. Afterwards, the resulting tissue supernatant is harvested, spun free of cells, and used in an ELISA or Luminex assay. For organoid assays, organoids are prepared in a monolayer format and treated with L. reuteri-conditioned medium or growth medium control as for the ex vivo tissue. Figure made with BioRender. Oxytocin measured by ELISA and normalized by tissue surface area secreted from ex vivo b) human and c) mouse intestinal tissue. d) oxytocin measured by Luminex secreted from uninduced and induced J2-NGN3 HIOs. e) oxytocin measured by ELISA secreted from induced J2-NGN3 HIOs. Point shape reflects unique patients (shown in triplicate) for human (b) or organoid batch (in triplicate) (d, e). For mouse (c), points represent unique mice and shape denotes sex (females as circles and males as squares). Significance values were determined from the least squares means derived from linear or linear mixed models with pairwise comparisons corrected using a Benjamini-Hochberg multiple testing correction (see Supplemental Tables S2 and S3). b: n = 5 patients for the duodenum through lower ileum and n = 4 patients for the colon regions with three replicate tissues each (12 or 15 datapoints total); c: n = 12 animals per region and condition; d: n = 3 HIO batches with two replicate monolayers per condition (6 datapoints total); e: n = 3 HIO batches with two replicate monolayers per condition (6 datapoints total).
