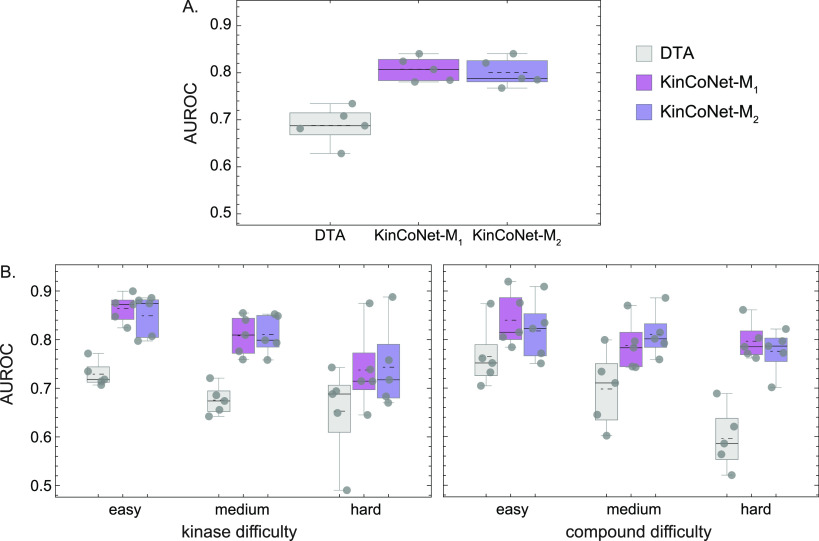
Figure 6.
Structure-based methods outperform structure-free methods for predicting whether a kinase-compound pair interacts. Comparison of the AUROC of DTC, KinCoNet-M1, and KinCoNet-M2 (A) on the overall test set across all kinase and compound difficulty levels, (B) on test sets with increasing kinase difficulty (left), and on test sets with increasing compound difficulty (right). Comparison of the correlation between experimental affinities and predictions from the three models. Each gray dot represents the AUC from a CV split. Black dotted lines represent the mean. DTA – structure-free models trained with the DTA architecture on DTC and PDBBind-kinase; KinCoNet-M1 – models trained on KinCo with docked structures selected by Autodock Vina combined with PDBBind-kinase; KinCoNet-M2 – models trained on KinCo with docked structures selected by M1 combined with PDBBind-kinase.