In this study, Zezulin et al. describe the essential role of transcription factor RUNX1 in controlling the inflammatory transcriptional program in granulocyte–monocyte progenitors and neutrophils. The loss of RUNX1 causes derepression of TLR4 and type I IFN signaling pathways and retroelements, resulting in neutrophil hyperresponsiveness and innate immunity disorders or malignancies.
Keywords: RUNX1, hematopoiesis, inflammation, retroelements, transposable elements
Abstract
The transcription factor RUNX1 is mutated in familial platelet disorder with associated myeloid malignancy (FPDMM) and in sporadic myelodysplastic syndrome and leukemia. RUNX1 was shown to regulate inflammation in multiple cell types. Here we show that RUNX1 is required in granulocyte–monocyte progenitors (GMPs) to epigenetically repress two inflammatory signaling pathways in neutrophils: Toll-like receptor 4 (TLR4) and type I interferon (IFN) signaling. RUNX1 loss in GMPs augments neutrophils’ inflammatory response to the TLR4 ligand lipopolysaccharide through increased expression of the TLR4 coreceptor CD14. RUNX1 binds Cd14 and other genes encoding proteins in the TLR4 and type I IFN signaling pathways whose chromatin accessibility increases when RUNX1 is deleted. Transcription factor footprints for the effectors of type I IFN signaling—the signal transducer and activator of transcription (STAT1::STAT2) and interferon regulatory factors (IRFs)—were enriched in chromatin that gained accessibility in both GMPs and neutrophils when RUNX1 was lost. STAT1::STAT2 and IRF motifs were also enriched in the chromatin of retrotransposons that were derepressed in RUNX1-deficient GMPs and neutrophils. We conclude that a major direct effect of RUNX1 loss in GMPs is the derepression of type I IFN and TLR4 signaling, resulting in a state of fixed maladaptive innate immunity.
The transcription factor (TF) RUNX1 is mutated in both sporadic and inherited forms of leukemia and myelodysplastic syndrome (MDS) (Bellissimo and Speck 2017). Inherited RUNX1 mutations cause familial platelet disorder with associated myeloid malignancy (FPDMM), which is characterized by thrombocytopenia, platelet activation defects, accelerated clonal hematopoiesis, and an up to 50% lifetime risk of leukemia (Churpek et al. 2015; Brown et al. 2020; Deuitch et al. 2021). The inflammatory disease eczema was reported in an FPDMM pedigree (Sorrell et al. 2012) and since then has been documented in up to 50% of families (Brown et al. 2020; Deuitch et al. 2021). FPDMM patients also have an increased incidence of asthma, reactive airway disease, rosacea, and allergies (Sacco et al. 2020). Chronic inflammation is a driver of leukemia (Kouroukli et al. 2022; Stubbins et al. 2022); therefore, inflammatory disorders in FPDMM patients may contribute to the elevated incidence of hematologic malignancies (Bellissimo and Speck 2017).
The regulation of inflammation by RUNX1 has been observed in multiple experimental contexts and occurs via diverse mechanisms. RUNX1 has been shown to promote, but more often to repress, inflammation. RUNX1 promotes inflammatory signaling in macrophages by binding the p50 subunit of nuclear factor κB (NF-κB), which augments the expression of several proinflammatory cytokine genes in response to Toll-like receptor 4 (TLR4) signaling (Luo et al. 2016). RUNX1 represses inflammation through its interactions with FOXP3, an essential transcription factor in regulatory T cells (Tregs) (Ono et al. 2007). RUNX1 represses inflammation in nonimmune lung alveolar epithelial cells in response to lipopolysaccharide (LPS)-induced lung injury by inhibiting IκB kinase (Tang et al. 2017).
RUNX1 also represses inflammation by negatively regulating type I interferon (IFN) production and signaling. Type I IFN signaling is triggered by type I IFN binding to the transmembrane IFNα/β receptor. The receptor dimerizes, bringing Janus-activated kinase 1 (JAK1) and tyrosine kinase 2 (TYK2) into close proximity, whereupon they phosphorylate each other, the cytoplasmic domains of the receptor subunits, and the signal transducer and activator of transcription (STAT) proteins STAT1 and STAT2. Phosphorylated STAT1 (p-STAT1), p-STAT2, and an IFN regulatory factor (IRF) assemble into a complex that translocates into the nucleus and activates the transcription of a group of IFN-stimulated genes (ISGs). Overexpression of dominant-negative forms of RUNX1 or RUNX1 knockdown induced the expression of type I IFNs and ISGs, while overexpressing RUNX1 had the opposite effect (DeKelver et al. 2014; Hu et al. 2022). Deletion of RUNX1 in resting B cells also increased the expression of ISGs and caused B cells to hyperrespond to LPS following prestimulation of the B cell receptor (Thomsen et al. 2021).
Type I IFN signaling is associated with the derepression of transposable elements (TEs). TEs consist of retrotransposons and DNA transposons, which together constitute almost 50% of the mammalian genome. Retrotransposons can be divided into two groups: the long terminal repeats and endogenous retroviruses (LTRs/ERVs) and non-LTR elements, including long or short interspersed nuclear elements (LINEs and SINEs). The chromatin accessibility of TEs is held in check by multiple epigenetic mechanisms including DNA methylation and histone modification, but in certain disease states TEs can become derepressed (Geis and Goff 2020). LTRs/ERVs, and in particular ERVK, often escape epigenetic silencing in both hematologic and solid malignancies (Matteucci et al. 2018; Kitsou et al. 2022), and dysregulated retrotransposon expression has been linked to inflammation in autoimmune and neurological diseases (Saleh et al. 2019). LTR/ERVs contain STAT1 binding sites and act as enhancers for ISGs (Chuong et al. 2016). In addition, bidirectional transcription of retrotransposons leads to the production of dsRNA, which can trigger the innate immune response by binding cytosolic RIG-I-like receptors that activate a signaling pathway, culminating in the expression of type I IFNs (Chen and Hur 2022). A study of 178 AML patients from the Cancer Genome Atlas (TCGA) found that patients with RUNX1 mutations had altered levels of transcripts from TEs, suggesting a role for RUNX1 in directly or indirectly regulating their expression (Colombo et al. 2018).
We previously showed that panhematopoietic RUNX1 deletion resulted in bone marrow (BM) neutrophils that secreted elevated amounts of tumor necrosis factor (TNF), macrophage inhibitory protein α (MIP-1α [or CCL3]), and IL-1α following activation of TLR4 signaling (Bellissimo et al. 2020). However, deleting RUNX1 in more differentiated neutrophils using a neutrophil-specific Cre (S100A8-Cre) did not affect the production of inflammatory molecules, indicating that RUNX1 protein was not functioning in neutrophils per se to regulate TLR4 signaling (Bellissimo et al. 2020). We hypothesized that instead RUNX1 was required in a neutrophil precursor to restrain inflammatory cytokine production by neutrophils. Single-cell RNA sequencing (scRNA-seq) analysis of RUNX1-deficient hematopoietic progenitors hinted that the dysregulation of inflammatory signaling occurred at the granulocyte–monocyte progenitor (GMP) stage (Bellissimo et al. 2020). Here we show that RUNX1 is required in GMPs to restrain inflammatory cytokine production by neutrophils. Loss of RUNX1 in GMPs up-regulates an inflammatory transcriptional program in both GMPs and neutrophils, characterized by an elevated type I IFN signature. Epigenetic profiling by assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq) and digital footprinting determined that the chromatin associated with both protein-coding genes and retrotransposons that gained accessibility upon RUNX1 loss was highly enriched for footprints for STAT1::STAT2 and multiple IRFs. RUNX1 occupies many genes encoding members of the type I IFN signaling pathway that are derepressed when RUNX1 is deleted, providing further evidence that RUNX1 directly regulates type I IFN signaling. Treatment of neutrophils with an inhibitor of type I IFN signaling ameliorated the hyperresponse of RUNX1-deficient neutrophils to LPS, whereas deletion of CD14 did not decrease the type I IFN signature, indicating that elevated type I IFN signaling augmented TLR4 signaling but not vice versa. Together, these data indicate that the hyperresponse of neutrophils to LPS-induced TLR4 signaling is a direct consequence of RUNX1's absence in GMPs, which elevates tonic type I IFN signaling.
Results
RUNX1 is required in GMPs to restrain inflammatory cytokine production by neutrophils
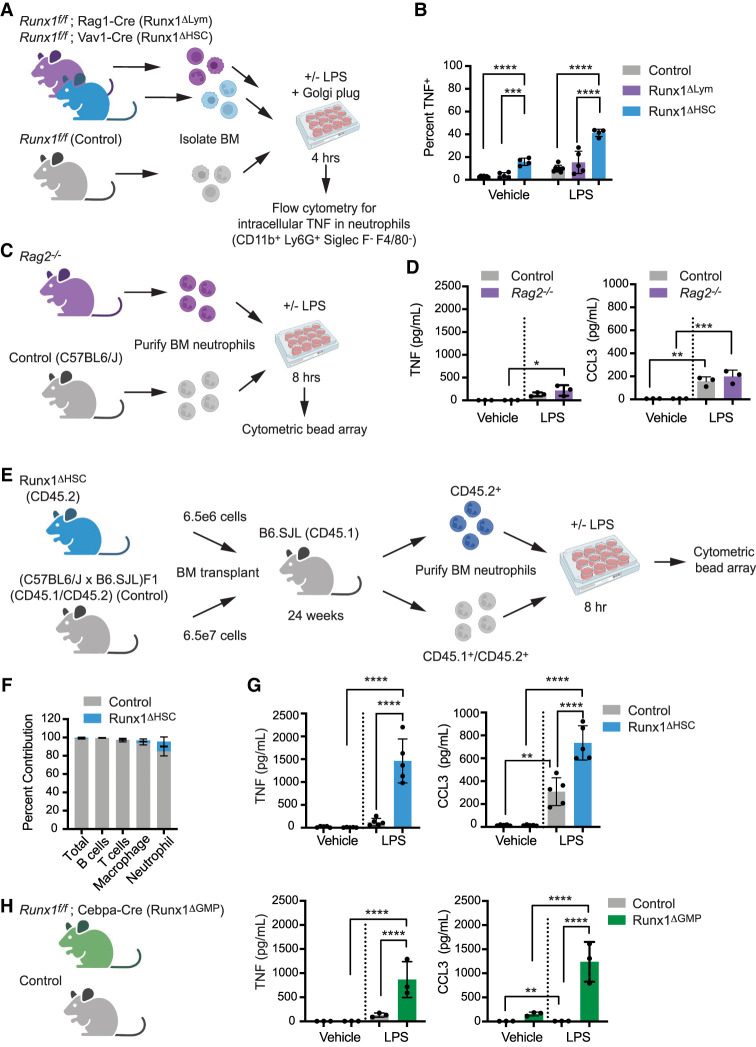
We used several complementary approaches to determine whether the hyperresponse to LPS was an intrinsic property of RUNX1-deficient neutrophils or an indirect effect of RUNX1 loss in another hematopoietic cell lineage. First, we addressed whether the aberrant neutrophil response was secondary to RUNX1 loss in lymphocytes (for example, loss of Tregs) by comparing the LPS response of neutrophils from mice in which RUNX1 was deleted in all hematopoietic cells by Vav1-Cre (referred to here as Runx1ΔHSC) with neutrophils from mice in which Runx1 was deleted only in lymphoid cells by Rag1-Cre (Runx1ΔLym) (Fig. 1A). We stimulated BM cells from Runx1ΔHSC, Runx1ΔLym, or control (Runx1f/f) mice ex vivo with vehicle or LPS for 4 h and then measured the percentage of TNF+ neutrophils by intracellular flow cytometry (see Supplemental Fig. S1A for gating strategy). The percentage of neutrophils that were TNF+ following LPS treatment was greater in Runx1ΔHSC compared with control BM cells but not in Runx1ΔLym BM, indicating that the loss of RUNX1 in lymphoid cells alone does not cause neutrophils to hyperrespond to LPS (Fig. 1B; Supplemental Fig. S1B). Since lymphocytes were not completely absent in Runx1ΔLym mice (Supplemental Fig. S1C,D), we also examined the inflammatory response of neutrophils from Rag2−/− mice that lack lymphocytes (Supplemental Fig. S1E,F; Shinkai et al. 1992). Purified neutrophils from Rag2−/− mice did not overproduce either TNF or CCL3, as determined using a cytometric bead array (CBA) (Fig. 1C,D), confirming that neutrophils from Runx1ΔHSC mice are not hyperresponsive because of lymphocyte defects.
Figure 1.
RUNX1 function in GMPs is necessary to restrict inflammatory cytokine production by neutrophils. (A) Schematic depicting an experiment to compare the effect of panhematopoietic (Runx1ΔHSC) versus lymphocyte-specific (Runx1ΔLym) RUNX1 loss on inflammatory cytokine production by neutrophils. BM cells were stimulated for 4 h ex vivo with vehicle or 100 ng/mL LPS. The percentage of neutrophils that were TNF+ was determined by flow cytometry. (B) Percentage of Runx1ΔHSC, Runx1ΔLym, and control BM neutrophils that were TNF+. Mean ± SD, one-way ANOVA, Tukey's multiple comparison test. Representative of three experiments. (C) Schematic depicting an experiment to examine the inflammatory phenotype of neutrophils from Rag2−/− mice. (D) Absolute quantification by CBA of inflammatory factors in the supernatant of neutrophils from control and Rag2−/− mice stimulated for 8 h with vehicle or 100 ng/mL LPS. Mean ± SD, one-way ANOVA plus Tukey's multiple comparison test. Representative of two experiments. (E) Schematic representation of the experimental design for generating BM chimeras by transplanting Runx1ΔHSC BM cells and a 10-fold excess of control BM cells into irradiated B6.SJL mice. Neutrophils derived from transplanted Runx1ΔHSC and control BM were purified by FACS 24 wk posttransplant and analyzed as in C and D. (F) Percentage of total BM cells, B cells (CD19+), T cells (CD3+), granulocytes (Gr1+CD11b+), and macrophages (CD11b+Gr1−) derived from Runx1ΔHSC versus control BM in transplant recipient mice. (G) Representative experiment of absolute quantification by CBA of inflammatory factors in the supernatant of control and Runx1ΔHSC neutrophils purified from transplant recipients and stimulated for 8 h with vehicle or 100 ng/mL LPS. Mean ± SD, one-way ANOVA plus Tukey's multiple comparison test. Representative of two experiments. (H) Absolute quantification by CBA of inflammatory factors in the supernatant of BM neutrophils from control or Runx1ΔGMP mice, analyzed as described in C. Mean ± SD, one-way ANOVA plus Tukey's multiple comparison test. Representative of nine experiments. For all experiments, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001.
To address whether other hematopoietic cells in the BM could be responsible for the hyperresponsive neutrophil phenotype, we generated BM chimeras by transplanting a 10:1 ratio of BM cells from control (C57BL6/J × B6.SJL) F1 and Runx1ΔHSC mice into irradiated B6.SJL mice (Fig. 1E). At 24 wk posttransplant, 100% of T and B cells and 97% of the neutrophils in recipient mice were derived from the transplanted control cells; thus, they far outnumbered the Runx1ΔHSC neutrophils (Fig. 1F). Runx1ΔHSC neutrophils purified from transplant recipient mice secreted more TNF and CCL3 than control neutrophils purified from the same recipient mice (Fig. 1G). Therefore, Runx1ΔHSC neutrophils in a BM consisting primarily of normal hematopoietic cells hyperrespond to LPS stimulation, consistent with a neutrophil-intrinsic defect.
In our previous scRNA-seq analysis of Runx1ΔHSC hematopoietic stem and progenitor cells, an aberrant inflammatory transcriptional signature was first detectable in GMPs (Bellissimo et al. 2020). To test whether RUNX1 loss in GMPs was sufficient to generate hyperresponsive neutrophils, we deleted RUNX1 using Cebpa-Cre (Runx1ΔGMP), which deletes primarily in GMPs (76%) and, to a lesser extent, in common lymphoid progenitors (9%), common myeloid progenitors (26%), and LSK cells (13%) (Wolfler et al. 2010). Cebpa-Cre efficiently deleted RUNX1 in GMPs and neutrophils (Supplemental Fig. S1G). BM neutrophils from Runx1ΔGMP mice overproduced TNF and CCL3 in response to LPS (Fig. 1H), demonstrating that loss of RUNX1 in GMPs is sufficient to establish a hyperresponsive neutrophil phenotype.
RUNX1 loss results in elevated levels of key TLR4 signaling molecules
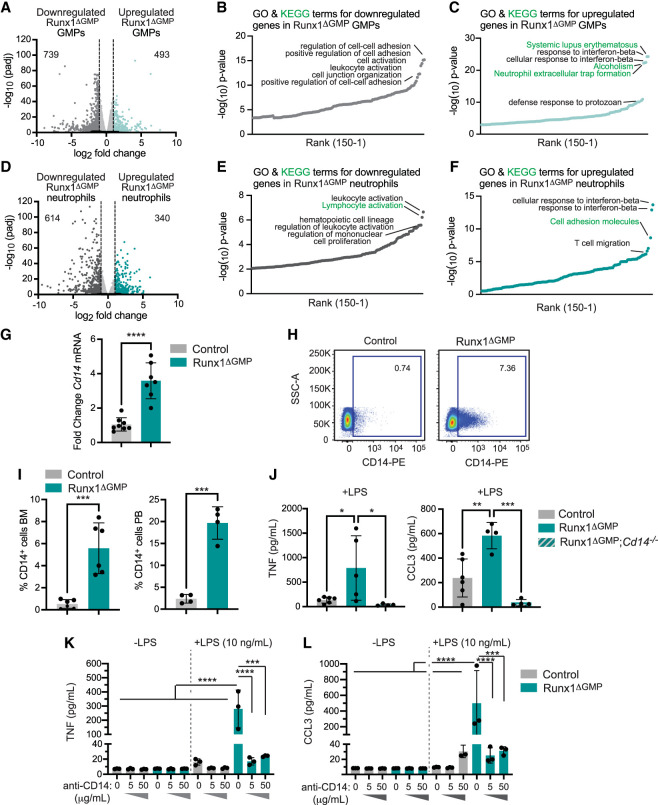
We analyzed the basal transcriptional changes (in the absence of LPS) in GMPs and neutrophils purified from Runx1ΔGMP mice by bulk RNA-seq (data quality is documented in Supplemental Fig. S2A,B). Approximately 1200 genes were differentially expressed in control versus Runx1ΔGMP GMPs and 954 genes were differentially expressed in control versus Runx1ΔGMP neutrophils, with more genes down-regulated than up-regulated in each cell type (Fig. 2A,D; Supplemental Table S2). Many enriched gene ontology (GO) terms for genes down-regulated in Runx1ΔGMP GMPs and neutrophils were related to cell adhesion and activation and regulation of leukocyte activation, respectively (Fig. 2B,E). GO terms for genes up-regulated in Runx1ΔGMP GMPs and neutrophils were related to inflammatory and immune responses, including “response to interferon β,” “defense response to protozoan,” and “neutrophil extracellular trap formation” (Fig. 2C,F).
Figure 2.
RUNX1 loss results in elevated levels of key TLR4 signaling molecules. (A) Volcano plot depicting global transcriptional changes between control and Runx1ΔGMP GMPs. Up-regulated genes with an adjusted P-value of <0.05 and a log2 fold change >1 are indicated by teal dots, and down-regulated genes with a log2 fold change less than −1 are indicated by dark-gray dots. The numbers of significantly up-regulated or down-regulated genes are indicated. (B) Top 150 enriched gene ontology (GO) (black text) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (green text) terms for genes down-regulated in Runx1ΔGMP GMPs. Enrichment of GO and KEGG terms was tested using Fisher's exact test (GeneSCF v1.1-p2). (C) Top 150 enriched GO and KEGG pathway terms for genes up-regulated in Runx1ΔGMP GMPs. (D) Volcano plot depicting global transcriptional changes between control and Runx1ΔGMP neutrophils. (E) Top 150 enriched GO and KEGG terms for genes down-regulated in Runx1ΔGMP neutrophils. (F) GO and KEGG terms for genes up-regulated in Runx1ΔGMP neutrophils. (G) RT-qPCR for Cd14 in Runx1ΔGMP neutrophils. Mean ± SD, unpaired, two-tailed Student's t-test. (H) Representative scatter plots of CD14 expression on BM neutrophils from control and Runx1ΔGMP mice. (I) Quantification of the percentage of CD14+ neutrophils in the BM (left) and PB (right) of control and Runx1ΔGMP mice. Mean ± SD, unpaired, two-tailed Student's t-test. Representative of 11 experiments, and a total of 32 mice were analyzed. (J) Absolute quantification by CBA demonstrating that deletion of Cd14 reduces TNF and CCL3 production by control and Runx1ΔGMP neutrophils in response to 10 ng/mL LPS. Mean ± SD, one-way ANOVA plus Tukey's multiple comparison test. Representative of two experiments, and a total of 24 mice were analyzed. (K) Absolute quantification by CBA demonstrating the effect of CD14-blocking antibody on TNF production by purified BM-derived neutrophils stimulated for 8 h with vehicle or a low dose of LPS (10 ng/mL). Mean ± SD, one-way ANOVA plus Tukey's multiple comparison test. Representative of two experiments and a total of eight mice were analyzed. (L) Effect of CD14-blocking antibody on CCL3 production by purified neutrophils stimulated for 8 h with vehicle or a low dose of LPS, as in K. For all panels, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001.
Expression of the Cd14 gene was up-regulated in Runx1ΔGMP neutrophils by RNA-seq and RT-qPCR (Fig. 2G). CD14 is a glycosylphosphatidylinositol-anchored TLR4 accessory protein present on the cell membrane and secreted in a soluble form that binds LPS and transfers it to TLR4 (Zanoni and Granucci 2013). The transfer of LPS from CD14 to TLR4 facilitates TLR4 endocytosis, activation of the TRIF pathway, and type I IFN production (Zanoni and Granucci 2013; Ciesielska et al. 2021). The percentages of CD14+ neutrophils in the BM and peripheral blood (PB) of Runx1ΔGMP mice were increased by ∼10 fold compared with control mice (Fig. 2H,I; Supplemental Fig. S3A). CD14 contributed to the hyperresponse, as neutrophils from Runx1ΔGMP mice deficient in CD14 produced lower levels of TNF and CCL3 in response to LPS (Fig. 2J). CD14 increases the sensitivity of TLR4 activation under conditions of low (picomolar) LPS concentrations but is not required for TLR4 responses to high LPS concentrations (Perera et al. 1997; Zanoni and Granucci 2013). Blocking antibodies against CD14 decreased the levels of TNF and CCL3 that were secreted into culture supernatants by Runx1ΔGMP neutrophils in response to a low concentration of LPS (10 ng/mL) (Fig. 2K,L) but not to a 10-fold higher concentration (100 ng/mL) (Supplemental Fig. S3B). In summary, elevated levels of CD14 contribute to the hyperresponse of Runx1ΔGMP neutrophils in response to low levels of LPS, and inhibiting CD14 could dampen the exaggerated response.
Loss of RUNX1 in GMPs increases the chromatin accessibility of genes involved in innate immune responses in both GMPs and neutrophils
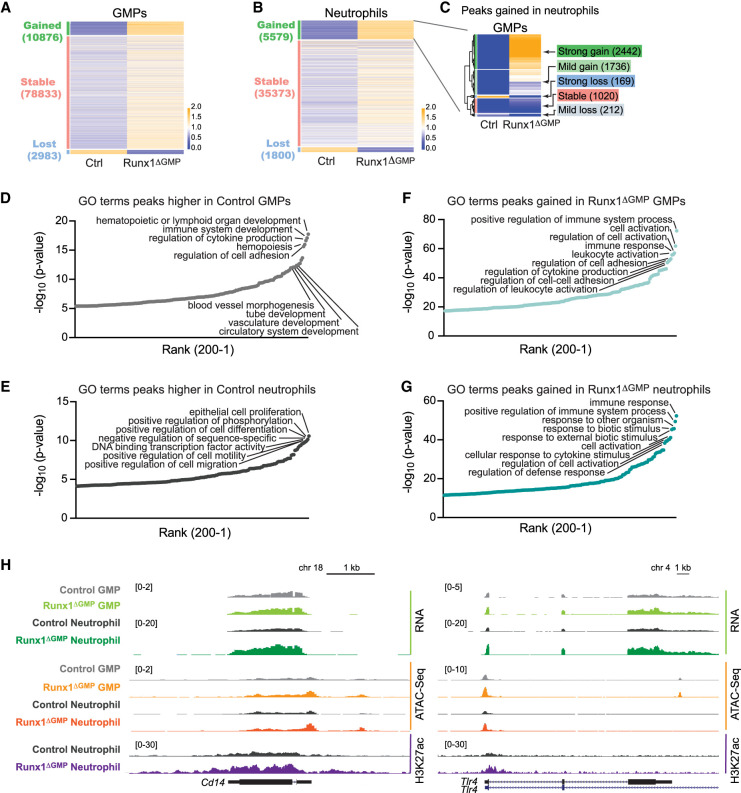
We hypothesized that epigenetic alterations in key inflammatory pathway genes are acquired in Runx1ΔGMP GMPs and propagated to neutrophils. To examine this, we performed ATAC-seq on non-LPS-stimulated GMPs and neutrophils. Overall, despite the fact that fewer genes were expressed in Runx1ΔGMP GMPs and neutrophils, more ATAC-seq peaks were gained than lost (Fig. 3A,B). Of the 5579 ATAC-seq peaks that were higher in Runx1ΔGMP neutrophils, 75% had been strongly (2442) or mildly (1736) gained in Runx1ΔGMP GMPs (Fig. 3C). Therefore, most of the increases in chromatin accessibility in Runx1ΔGMP neutrophils originated in Runx1ΔGMP GMPs. In contrast, only 36% of peaks lost in Runx1ΔGMP neutrophils had been lost in GMPs (Supplemental Fig. S4A). Many GO terms associated with peaks lost in Runx1ΔGMP GMPs or neutrophils (i.e., associated with peaks higher in control GMPs and neutrophils) are related to developmental or cell biological processes (Fig. 3D,E). On the other hand, terms associated with peaks gained in Runx1ΔGMP GMPs or neutrophils (Fig. 3F,G) or gained peaks shared by Runx1ΔGMP GMPs and neutrophils (Supplemental Fig. S4B; Supplemental Table S3) were related to immune responses, suggesting that RUNX1 loss increased the accessibility of genes associated with immune cell activation at the GMP stage, including Tlr4 and Cd14 (Fig. 3H). The expression of genes proximal to the lost and gained peaks exhibited small but significant changes in expression correlating with the changes in chromatin accessibility (Supplemental Fig. S4C).
Figure 3.
Loss of RUNX1 increases the chromatin accessibility of genes involved in innate immune responses in GMPs and neutrophils. (A) Heat map of ATAC-seq signals for control (Ctrl) and Runx1ΔGMP GMPs. Peaks are categorized as gained, lost (in Runx1ΔGMP GMPs), or stable (RPKM fold change <2). The color represents the relative RPKM and the mean RPKM of control and Runx1ΔGMP cells. (B) Heat map of ATAC-seq signals in control and Runx1ΔGMP neutrophils. (C) Heat map of ATAC-seq signal in control and Runx1ΔGMP GMPs for peaks gained in Runx1ΔGMP neutrophils. The peaks were clustered hierarchically and then segregated into five groups using the cutree function in R. (D) Enriched GO biological terms for peaks higher in control GMPs (i.e., peaks lost in Runx1ΔGMP GMPs). The top 200 GO biological terms are plotted. (E) Enriched GO biological terms for peaks higher in control neutrophils (i.e., peaks lost in Runx1ΔGMP neutrophils). (F) Enriched GO biological terms for peaks gained in Runx1ΔGMP GMPs. (G) Enriched GO biological terms for peaks gained in Runx1ΔGMP neutrophils. (H) Genome browser view showing normalized RNA-seq, ATAC-seq, and H3K27ac signals for the Tlr4 and Cd14 genes in control and Runx1ΔGMP GMPs and neutrophils.
ChIP-seq for the active enhancer mark H3K27ac found that gained peaks in Runx1ΔGMP neutrophils were also enriched for GO terms related to innate immune responses (Supplemental Fig. S4D,E), confirming that the enhancers of these genes were in a more active state. We examined how H3K27ac peaks gained and lost in Runx1ΔGMP neutrophils were altered in control neutrophils following LPS stimulation to determine their relationship to the LPS response. Of the 850 peaks lost in Runx1ΔGMP neutrophils in the absence of LPS (i.e., higher in control neutrophils) (Supplemental Fig. S4D, light-teal box), 677 (80%) were lower in control neutrophils following LPS stimulation (Supplemental Fig. S4D, dark-teal box) relative to control neutrophils + vehicle (Supplemental Fig. S4D,F, light-teal box). Of the 1056 peaks gained in Runx1ΔGMP neutrophils relative to control neutrophils in the absence of LPS (Supplemental Fig. S4D, lavender box), 698 (66%) were higher in control neutrophils following LPS stimulation (Supplemental Fig. S4D, dark-purple box) compared with control neutrophils + vehicle (Supplemental Fig. S4D,F, pink box). Therefore, most of the lost and gained peaks in Runx1ΔGMP neutrophils in the absence of LPS were related to the response of control neutrophils to LPS, suggesting that Runx1ΔGMP neutrophils are primed to respond to LPS.
RUNX1 restrains tonic type I IFN signaling
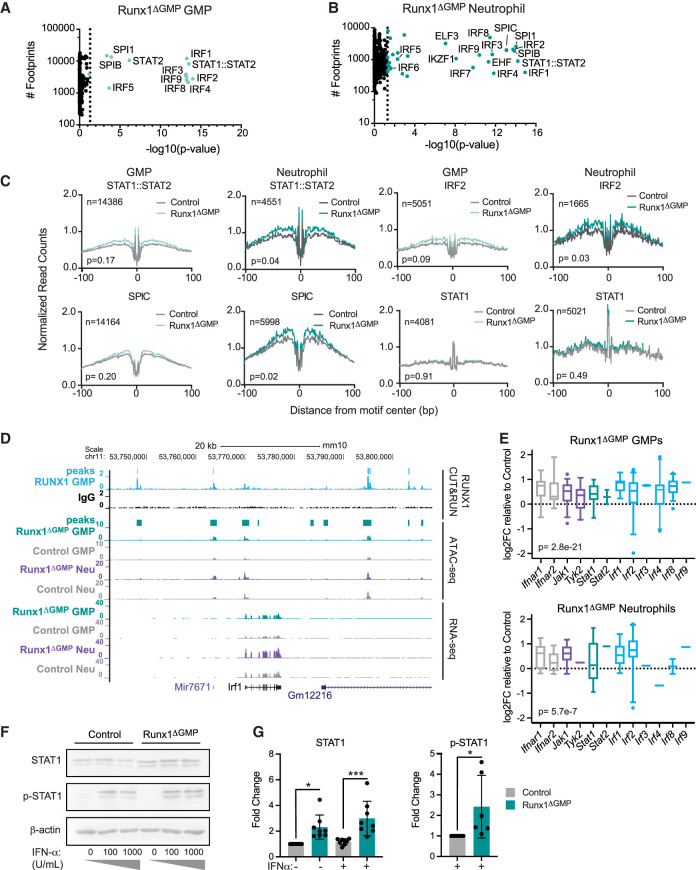
We inferred which TFs were responsible for opening the chromatin following RUNX1 loss by digital footprinting (Li et al. 2019). Digital “footprints” in ATAC-seq data result from the Tn5 enzyme's inability to cleave where TFs are bound to DNA, which results in a dip in reads or “footprint,” in peaks that can then be matched to TF motifs to infer the specific TF bound. As expected, footprints of all three RUNX TFs (RUNX1, RUNX2, and RUNX3 bind the same motifs) and multiple GATA TFs were highly enriched in peaks that were lost in GMPs when RUNX1 was deleted (Supplemental Fig. S5A; Supplemental Table S4). Footprints for RUNX TFs were also highly enriched in regions of decreased accessibility in Runx1ΔGMP neutrophils, as were footprints for several other TFs (e.g., Bhlha15, FOXB1, and ZSCAN4), though they were less enriched than RUNX footprints (Supplemental Fig. S5A).
The most highly enriched TF footprints in ATAC-seq peaks gained in both Runx1ΔGMP GMPs and neutrophils were for multiple IRFs and STAT1::STAT2 (Fig. 4A,B). Footprints for several E-twenty-six (ETS) TFs, SPI1 (PU.1), SPIB, and SPIC were also enriched in Runx1ΔGMP neutrophils (Fig. 4B). The chromatin surrounding the STAT1::STAT2 (Fig. 4C), IRF2 (Fig. 4C), IRF3, IRF4, IRF9, and SPIC footprints (Supplemental Table S4) was significantly more accessible in Runx1ΔGMP neutrophils, indicating that these TFs were opening the chromatin around their bound sites.
Figure 4.
Chromatin opened following RUNX1 loss is enriched for footprints of TF effectors of type I IFN signaling. (A) Scatter plots showing enriched digital TF footprints at regions of chromatin with increased accessibility in Runx1ΔGMP GMPs relative to controls. The number of footprints for each TF at regions of chromatin with increased accessibility is displayed on the Y-axis for the Runx1ΔGMPcells. Colored circles indicate P < 0.05; P-value was calculated using biFET. (B) Scatter plots showing enriched digital TF footprints at regions of chromatin with increased accessibility in Runx1ΔGMP neutrophils relative to controls as in A. (C) Footprint profile plots for selected TFs showing average normalized read counts and P-values calculated by HINT-differential (Li et al. 2019) using all peaks in control and Runx1ΔGMP cells. (D) Genome browser view showing RUNX1 occupancy determined by CUT&RUN, normalized ATAC-seq, and RNA-seq for the Irf1 gene in control and Runx1ΔGMP GMPs and neutrophils. Merged peaks for RUNX1 and ATAC-seq are indicated above the tracks for GMPs. (E) Box and whisker plots of log2FC in ATAC-seq signal relative to control for type I IFN pathway genes in Runx1ΔGMP GMPs and neutrophils. Whiskers represent the fifth to 95th percentile. (F) Western blot for STAT1 and phosphorylated (p) STAT1 plus β-actin control in control and Runx1ΔGMP neutrophils in the presence or absence of IFN-α. (G) Summary of Western blots in F. Mean ± SD. Dots indicate lanes quantified using ImageJ. ANOVA plus Tukey's multiple comparison test for STAT1; two-tailed, unpaired t-test for p-STAT1. Representative of eight experiments, and a total of 16 mice were analyzed. (*) P ≤ 0.05, (***) P ≤ 0.001.
IRFs and STAT1::STAT2 are downstream effectors of type I IFN signaling; thus, enrichment of their footprints suggests that type I IFN signaling was elevated in Runx1ΔGMP neutrophils and GMPs. In contrast, STAT1 footprints were not enriched in opened chromatin, and the accessibility of chromatin around STAT1 footprints was not significantly increased in either Runx1ΔGMP GMPs or neutrophils (Fig. 4A–C). Therefore, we infer that type II IFN signaling, which is mediated by the binding of STAT1 dimers to GAS sites, is not elevated to the same extent as type I IFN signaling. STAT3 and STAT5 footprints were also not enriched in gained peaks in Runx1ΔGMP GMPs and neutrophils, and the chromatin accessibility around their footprints was not increased (Fig. 4A; Supplemental Fig. S5B), suggesting that cytokine signaling through the JAK/STAT pathway was not elevated. Footprints for NF-κB TFs (NFKB1, NFKB2, REL, and RELA) were also not enriched (Fig. 4A; Supplemental Fig. S5B), indicating that TLR4 signaling through the NF-κB arm of the pathway was not activated in the absence of LPS stimulation. In conclusion, RUNX1 loss predominantly increased chromatin accessibility of IRF- and STAT1::STAT2-occupied sites in both GMPs and neutrophils, suggesting that type I IFN signaling was elevated in both GMPs and neutrophils. These results are consistent with the RNA-seq data showing up-regulation of genes associated with “cellular response to interferon-β” and “response to interferon-β” in Runx1ΔGMP neutrophils and GMPs.
We considered why Runx1ΔGMP GMPs and neutrophils had acquired a type I IFN signature, which is generally associated with autoimmune diseases (Tanaka et al. 2022). One possibility is that type I IFNs are produced at higher levels by Runx1ΔGMP cells. However, we found no evidence for increased production of IFN-α and/or IFN-β by neutrophils using CBA assays or increased levels of IFN-α and/or IFN-β in the PB or BM with high-sensitivity ELISA assays. Furthermore, expression from most of the Ifna and the Ifnab genes was undetectable in GMPs and neutrophils by RNA-seq. Therefore, increased type I IFN levels are unlikely to be the cause of increased IRF and STAT1::STAT2 occupancy, though we cannot rule this out since low picomolar levels of IFN are sufficient to activate signaling (Yarilina et al. 2008). Another possibility is that RUNX1 directly or indirectly represses the expression of signaling molecules in the type I IFN pathway to activate constitutive IFN-independent signaling (Wang et al. 2017; Michalska et al. 2018). To examine whether RUNX1 binds to and potentially directly regulates type I IFN pathway genes, we mapped RUNX1 occupancy by cleavage under targets and release using nuclease (CUT&RUN) in GMPs (Skene and Henikoff 2017). We found that RUNX1 occupies multiple genes encoding proteins in the core type I IFN signaling pathway, including receptors (Ifnar1 and Ifn1r2), an associated kinase (Tyk2), and transcription factor effectors (Stat1, Stat2, Irf1, Irf2, Irf3, Irf4, Irf7, Irf8, and Irf9) (Fig. 4D; Supplemental Table S5). Deletion of RUNX1 in GMPs increased the chromatin accessibility of all these genes in GMPs and a subset of these genes in neutrophils (Fig. 4E). Furthermore, STAT1 protein levels were modestly increased in Runx1ΔGMP neutrophils (Fig 4F,G; Supplemental Fig. S6A–D), and RNA-seq read counts for type I IFN receptors (Ifnar1 and Ifnar2) and two IRFs (Irf1 and Irf9) were significantly higher in Runx1ΔGMP GMPs (Supplemental Fig. S6E). We conclude that RUNX1 may directly restrain the expression of multiple genes encoding proteins in the type I IFN signaling pathway.
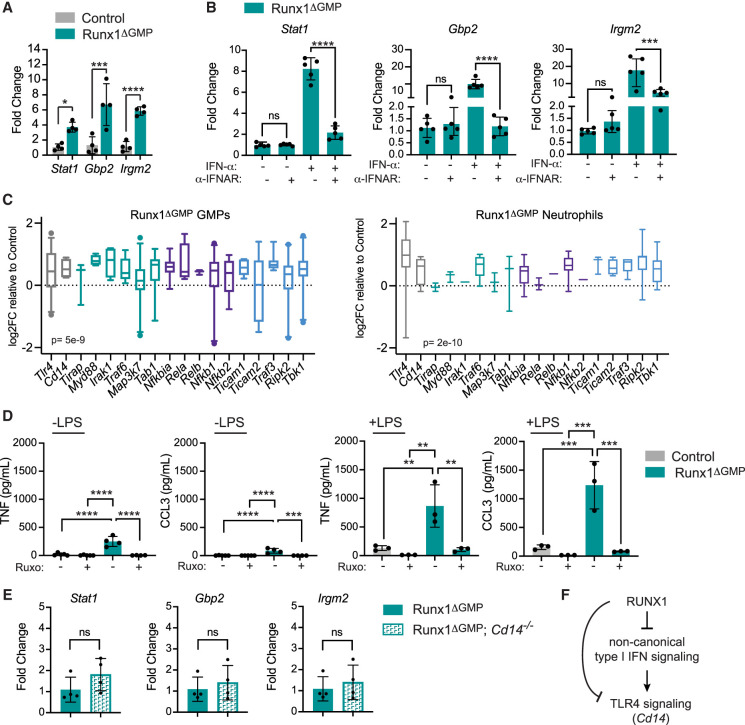
The failure to detect type I IFNs has been reported in other studies of cells with transcriptional and epigenetic type I IFN signatures (Michalska et al. 2018; Espinet et al. 2021). In some cases, it was shown that basal ISG expression is activated by IFN-independent noncanonical/tonic type I IFN signaling mediated by unphosphorylated (U) STATs (U-STAT1 and U-STAT2) complexed with IRF1 or IRF9 (Wang et al. 2017; Michalska et al. 2018; Platanitis et al. 2019). Unlike the canonical, IFN-stimulated pathway, IFN-independent tonic type I IFN signaling does not require activation of the type I IFN receptor (Wang et al. 2017). To determine whether the elevated basal expression of ISGs (Stat1, Gbp2, and Irgm2) in Runx1ΔGMP neutrophils (Fig. 5A) could be due to IFN-independent noncanonical signaling, we examined whether blocking the activity of IFNAR could reduce the levels of ISG mRNAs in Runx1ΔGMP neutrophils in the absence of IFN-α stimulation. A blocking antibody to IFNAR did not significantly decrease the basal levels of Stat1, Gbp2, or Irgm2 mRNAs in Runx1ΔGMP neutrophils in the absence of IFN-α, whereas it potently inhibited activation of the IFN-α-stimulated canonical pathway and expression of ISGs (Fig. 5B). We conclude that the increased expression of ISGs in Runx1ΔGMP neutrophils is caused by elevated tonic type I IFN-induced signaling due to increased expression of multiple proteins in the type I IFN signaling pathway. This either elevates IFN-independent signaling or sensitizes the neutrophil's response to undetectably low levels of type I IFN.
Figure 5.
RUNX1 restrains tonic type I IFN signaling. (A) RT-qPCR demonstrating baseline expression of three IGSs in control and Runx1ΔGMP neutrophils. Mean ± SD, two-tailed, unpaired t-test. Representative of two experiments, and total of eight mice were analyzed. (B) A blocking antibody against the type I IFN receptor (α-IFNAR) reduces type I IFN signaling through the canonical pathway (+IFN-α) but not tonic signaling through a noncanonical pathway (−IFN-α), as measured by the expression of ISGs by RT-qPCR. All data are from Runx1ΔGMP neutrophils. Representative of two experiments, and total of 12 mice were analyzed. Mean ± SD, ANOVA plus Tukey's multiple comparison test. (C) Box and whisker plots of log2FC in ATAC-seq signal relative to control for TLR4 pathway genes in Runx1ΔGMP GMPs and neutrophils. Whiskers represent the fifth to 95th percentile. ATAC-seq peaks were assigned to genes by genomic region enrichment of annotations tool (GREAT) (McLean et al. 2010). (D) Ruxolitinib (20 μM) decreases the production of TNF and CCL3 by neutrophils in response to LPS. Representative of two experiments, and total of 15 mice were analyzed. Mean ± SD, ANOVA plus Tukey's multiple comparison test. For all figures, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001, (ns) not significant. (E) The type I IFN signature is not caused by elevated levels of the TLR4 coreceptor CD14. RT-qPCR demonstrating baseline expression of three IGSs in Runx1ΔGMP and Runx1ΔGMP;Cd14−/− neutrophils. Representative of two experiments, and total of 14 mice were analyzed. Mean ± SD, two-tailed, unpaired t-test. (F) Schematic diagram to summarize the relationship between RUNX1, type I interferon, and TLR4 signaling.
RUNX1 occupies multiple genes encoding proteins in the TLR4 signaling pathway as well, including the core signaling components (Cd14, Irak2, Irak3, Ripk2, Tab2, Tbk1, Ticam1, Traf3, and Traf6) and key transcription factor effectors (Irf3, Nfkb1, Nfkb2, Nfkbia, Rela, and Relb) (Supplemental Table S5). Deletion of RUNX1 in GMPs significantly increased the chromatin accessibility of all these genes in GMPs and the majority of these genes in neutrophils (Fig. 5C), suggesting that RUNX1 may also directly regulate TLR4 signaling.
Finally, we examined the cross-talk between the type I IFN and TLR4 pathways. To determine whether elevated type I IFN signaling contributed to the hyperresponse to LPS, we stimulated Runx1ΔGMP neutrophils with LPS and tested whether the JAK1/JAK2 inhibitor ruxolitinib (Ruxo) decreased the production of TNF and CCL3. TNF and CCL3 levels were significantly decreased by ruxolitinib, indicating that type I IFN signaling contributes to the overly exuberant response of Runx1ΔGMP neutrophils to LPS (Fig. 5D). We next asked whether elevated levels of the TLR4 coreceptor CD14 was responsible for the interferon signature. Runx1ΔGMP neutrophils lacking the TLR4 coreceptor CD14 expressed the same levels of ISGs compared with neutrophils from Runx1ΔGMP mice (Fig. 5E), indicating the type I IFN signature was not dependent on TLR4 pathway activation. Together, the data suggest that RUNX1 directly restrains type I IFN and TLR4 signaling by binding and repressing multiple genes encoding proteins in their core pathways. Type I IFN signaling also augments TLR4 signaling, but TLR4 signaling does not contribute to the type I IFN signature (Fig. 5F).
RUNX1 restrains the chromatin accessibility and expression of retroelements
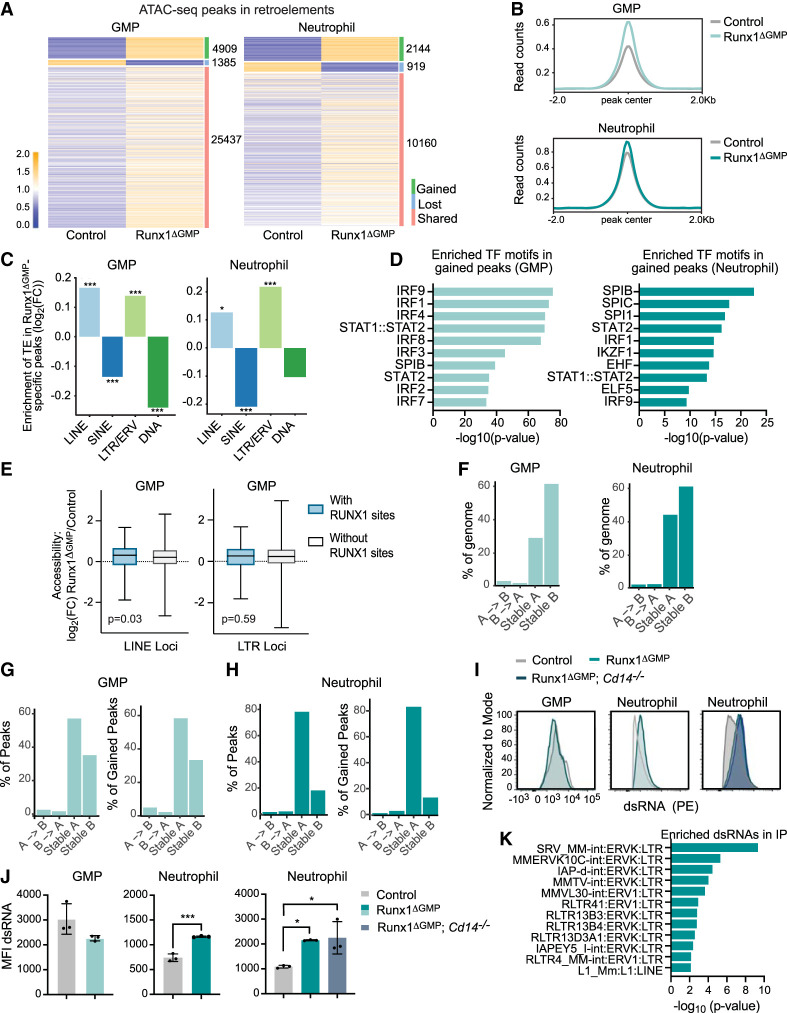
Elevated tonic type I IFN signaling has been associated with the expression of retroelements, including long terminal repeats/endogenous retroviruses (LTRs/ERVs) and long interspersed nuclear elements (LINEs) (Gázquez-Gutierréz et al. 2021). We examined chromatin accessibility using ATAC-seq data from Runx1ΔGMP GMPs and neutrophils and found an overall increase in the chromatin accessibility of transposable elements (TEs) (Fig. 6A,B). The gained ATAC-seq peaks in Runx1ΔGMP GMPs and neutrophils were enriched for LINEs (LINE/L1) and LTR/ERV retroelements, while short interspersed nuclear elements (SINEs) and DNA transposons were significantly underrepresented in gained peaks in one or both Runx1ΔGMP cell types (Fig. 6C). Gained ATAC-seq peaks in retroelements were enriched for many of the TF motifs identified in the genome-wide TF footprinting analysis (Figs. 4A,B, 6D), including motifs for multiple IRFs, STAT1::STAT2, and SPIB, suggesting that the increased chromatin accessibility of LINEs and LTR/ERV retroelements in Runx1ΔGMP GMPs and neutrophils was caused by elevated tonic type I IFN signaling. We also examined whether RUNX1 could play a direct role in repressing the chromatin accessibility of LINE and LTR/ERV retroelements in GMPs by comparing the ATAC-seq signal in retroelements with and without RUNX1 binding sites. The accessibility of LINEs bound by RUNX1 was significantly higher in Runx1ΔGMP GMPs compared with LINEs not bound by RUNX1 (Fig. 6E), though the difference in accessibility was small. There was no difference in the accessibility of LTR/ERV elements bound or not bound by RUNX1 in Runx1ΔGMP GMPs. We conclude that RUNX1 may directly constrain the chromatin accessibility of LINEs in GMPs but not that of LTRs/ERVs.
Figure 6.
Loss of RUNX1 in GMPs derepresses TEs. (A) Heat maps of ATAC-seq peaks in TEs in GMPs and neutrophils. The number of gained, lost, and shared peaks is listed at the right of each heat map. (B) Averaged ATAC-seq peak profile plots (normalized to bins per million mapped reads [BPM]) in TEs in control and Runx1ΔGMP neutrophils and GMPs. (C) Enrichment or depletion of the different classes of TEs (LINEs, SINEs, LTRs/ERVs, or DNA) in gained ATAC-seq peaks in Runx1ΔGMP neutrophils and GMPs. The Y-axis represents the log2 fold change of the number of Runx1ΔGMP-specific peaks overlapping with TEs over the median number of the randomly selected peaks overlapping with TEs. Positive log2 fold change = enrichment, and negative log2 fold change = depletion. (D) Top 10 TF binding motifs enriched in Runx1ΔGMP neutrophil-specific or Runx1ΔGMP GMP-specific ATAC–seq peaks that overlapped with TEs. (E) Box and whisker plots of log2FC in ATAC-seq signal relative to control for LINE or LTR loci with and without RUNX1 binding sites in control GMPs. (F) Genome-wide redistribution of chromatin compartments A and B in Runx1ΔGMP GMPs (left panel) and neutrophils (right panel) from control. (G) Percentage of all (left panel) and gained (right panel) ATAC-seq peaks in chromatin compartments A/B of GMPs. (H) Percentage of all (left panel) and gained (right panel) ATAC-seq peaks in chromatin compartments A/B of neutrophils. (I) Representative histograms for the mean fluorescence intensities (MFI) of dsRNA in the dsRNA+ neutrophils or GMPs. (J) Quantification of the relative MFI of dsRNA in the dsRNA+ neutrophils or GMPs. Statistics represent two-tailed unpaired Student's t-tests. Representative of two experiments, and total of 12 mice were analyzed. (*) P ≤ 0.05, (***) P ≤ 0.001. (K) IP of dsRNA with the 9D5 antibody followed by RNA-seq in Runx1ΔGMP and control neutrophils. Graph shows the dsRNA species detected and enriched in the IP from Runx1ΔGMP neutrophils over control neutrophils.
TEs contribute to higher-order chromatin structure (Lawson et al. 2023). To determine whether the increased accessibility of retroelements correlated with the repartitioning of chromatin between the euchromatic A and heterochromatic B compartments, we performed Hi-C (data quality documented in Supplemental Fig. S7A). Overall, RUNX1 deficiency had relatively little effect on genome compartmentalization, as <4% of chromatin shifted from the A to B or the B to A compartments in either GMPs or neutrophils (Fig. 6F). We also assessed whether ATAC-seq peaks in LINEs and LTR/ERV retroelements were enriched in chromatin that shifted between the A and B compartments. ATAC-seq peaks in retroelements and peaks gained in retroelements were most enriched in the stable A compartment, especially in Runx1ΔGMP neutrophils (Fig. 6G,H; Supplemental Fig. S7B,C). Less than 4% of overall and gained ATAC-seq peaks in retroelements in Runx1ΔGMP GMPs and neutrophils shifted between the A and B compartments. In summary, loss of RUNX1 does not globally reorganize the chromatin or the distribution of retroelements between the A and B compartments in Runx1ΔGMP GMPs and neutrophils.
Retroelements are a source of dsRNA, and excess dsRNA is known to activate cytoplasmic dsRNA sensors that trigger the production of type I IFNs (Chen and Hur 2022). To determine whether increased chromatin accessibility in RUNX1-deficient cells resulted in the overproduction of dsRNA, we performed flow cytometry using a dsRNA antibody (Son et al. 2015). The percentage of dsRNA+ cells and the median fluorescence intensity (MFI) of dsRNA per cell were increased in Runx1ΔGMP neutrophils but not in GMPs (Fig. 6I,J); therefore, dsRNA, which is a potent inducer of type I IFN (Michalska et al. 2018), is not responsible for the interferon signature in GMPs. The increased level of dsRNA in Runx1ΔGMP neutrophils was not caused by elevated tonic TLR4 signaling, as deletion of the TLR4 coreceptor CD14 did not reduce it (Fig. 6I,J). We immunoprecipitated and sequenced dsRNA from neutrophils with the same antibody used for flow cytometry to determine which dsRNAs were overexpressed. Immunoprecipitants of dsRNA from Runx1ΔGMP neutrophils were enriched for multiple LTRs/ERVKs and one LINE subfamily (Fig. 6K), while other ERV subfamilies and dsRNA from DNA transposons were significantly depleted (Supplemental Table S6). In conclusion, loss of RUNX1 elevates tonic type I IFN signaling and increases the chromatin accessibility of LINEs and LTR/ERV retroelements in GMPs and neutrophils and the production of dsRNA from a subset of LTRs/ERVKs and LINEs in neutrophils.
Haploinsufficiency of RUNX1 may alter the properties of mouse and human neutrophils
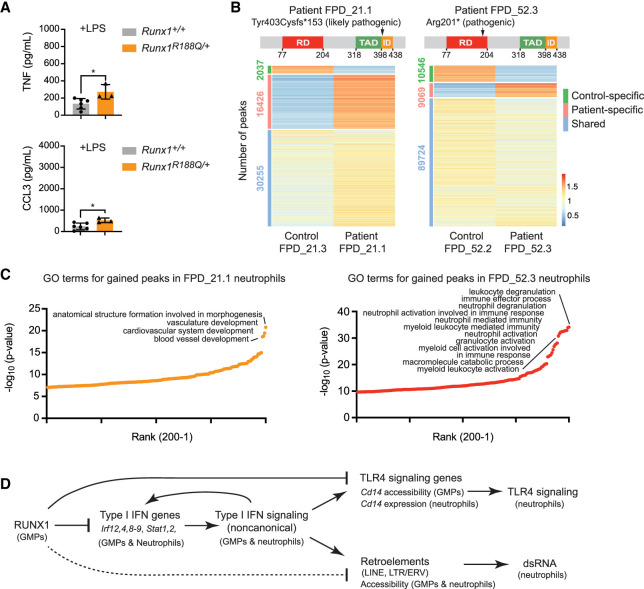
FPDMM is caused by germline monoallelic RUNX1 mutations (Song et al. 1999). To determine whether a monoallelic Runx1 mutation is sufficient to alter inflammatory cytokine production, we analyzed neutrophils from mice heterozygous for an R188Q mutation that affects a DNA-contacting residue (equivalent to R201Q in humans). The Runx1R188Q mutation caused a small but significant increase in the amount of TNF and CCL3 produced by neutrophils in response to LPS, demonstrating that monoallelic Runx1 mutations are sufficient to cause a mildly hyperresponsive neutrophil phenotype (Fig. 7A). We also analyzed peripheral blood neutrophils from two patients with FPDMM and two unaffected family members by ATAC-seq. Neutrophils from each FPDMM patient and their family members were isolated and processed in parallel. FPD_21.1, a 9-yr-old boy, had a frameshift mutation (Tyr403Cysfs*153) in what is labeled as the transactivation domain by Clinical Genome Resource (ClinGen) (Fig. 7B; Homan et al. 2021), though functional transactivation assays identified it as an inhibitory domain (Kanno et al. 1998). The Tyr403Cysfs*153 mutation is classified as likely pathogenic by ClinGen (Homan et al. 2021). Patient FPD_21.1 had a normal complete blood count (CBC) except for low platelets (73 K/µL), elevated eosinophils (5.7%), and a small increase in immature granulocytes (0.6%), as well as a persistent history of eczema. No known members of the FPD_21 family have had leukemia. FPD_52.3, an adult male, had a pathogenic nonsense mutation (Arg201*) in the DNA binding RUNT domain, resulting in a nonfunctional protein (Fig. 7B). Patient FPD_52.3 also had a persistent history of eczema and environmental allergies. His CBCs were within normal ranges except for platelets (94 K/µL) and eosinophils (14.1%). Three members of the FPD_52 family developed leukemia.
Figure 7.
Haploinsufficiency of RUNX1 may alter the properties of mouse and human neutrophils. (A) Absolute quantification by CBA of TNF and CCL3 in the supernatant of FACS-purified BM neutrophils from wild-type or Runx1R188Q/+ mice stimulated with 10 ng/mL LPS. Mean ± SD, two-tailed, unpaired t-test. Representative of two experiments, and a total of nine mice were analyzed. (*) P ≤ 0.05. (B, top) Schematic diagram showing the location of mutations in the RUNX1 protein in patients FPD_21.1 and FPD_52.3. ClinGen classifications of the mutations are indicated. (RD) DNA-binding RUNT domain, (TAD) transactivation domain, (ID) inhibitory domain as defined in functional assays (Kagoshima et al. 1993; Kanno et al. 1998). (Bottom) Heat maps of ATAC-seq signals for the two FPDMM patients and nonaffected family members (control). (C) Enriched GO terms for patient-specific peaks. The top 200 GO terms are plotted. (D) Model of the inflammatory regulatory network in GMPs and neutrophils directly and indirectly regulated by RUNX1. The dotted line indicates that the regulation of retroelement chromatin accessibility by RUNX1 may be primarily indirect.
The ratio of gained/lost peaks differed in neutrophils from FPD_21.1 and FPD_52.3 compared with their unaffected family members (Fig. 7B), as did the enriched GO biological terms (Fig. 7C). The most highly enriched terms associated with peaks higher in FPD_21.1 than in his unaffected father (FPD_21.3) were related to vascular development, while those higher in FPD_52.3 relative to his unaffected wife (FPD_52.2) were related to neutrophil activation, suggesting that FPD_52.3 neutrophils were in a more activated state (Fig. 7C). Terms related to type I IFN responses were not enriched in either patient or in control neutrophils. Many factors including age, environment, genetic background, health status, the nature of the RUNX1 mutations, and the partial penetrance of inflammatory conditions in FPDMM patients could account for the differences between mice and patients and between the two patients.
Discussion
We previously reported that panhematopoietic RUNX1 loss resulted in neutrophils that hyperrespond to LPS, but that RUNX1 does not function in neutrophils per se to restrain the response (Bellissimo et al. 2020). Here we demonstrate that RUNX1 is required in a neutrophil precursor, either in the GMP or downstream from the GMP, to restrain a proinflammatory epigenetic and transcriptional program in neutrophils. The proinflammatory program established in GMPs when RUNX1 was lost is characterized by a type I IFN signature and the derepression of retroelements, both of which result from elevated tonic type I IFN signaling (Fig. 7D). RUNX1 binds multiple genes encoding proteins in the type I IFN signaling pathway, and we hypothesize that a direct consequence of RUNX1 loss is the derepression of one or more of these genes. Interferon-stimulated genes include several that encode core components of the type I IFN signaling pathway; therefore, a positive feedback loop would be established as a result of RUNX1 loss to sustain tonic type I IFN signaling (Michalska et al. 2018). Elevated tonic type I IFN signaling also sensitizes the TLR4 signaling pathway and contributes to the overproduction of inflammatory cytokines by neutrophils in response to activation of TLR4 signaling (Fig. 7D).
The epigenetic rewiring in GMPs and neutrophils that results from RUNX1 loss has mechanistic parallels to trained immunity (Kalafati et al. 2022). In trained immunity, exposure to a pathogen induces epigenetic alterations in hematopoietic progenitors that are propagated to differentiated innate immune myeloid cells, causing them to respond more robustly to the same pathogen in a later encounter (Kalafati et al. 2022). This aspect of trained immunity provides obvious benefits to the infected host. Mechanistic parallels between trained immunity and RUNX1 loss include (1) epigenetic and/or transcriptional changes in GMPs, (2) acquisition of a type I IFN signature, and (3) neutrophils that hyperrespond to TLR ligands. For example, treatment of mice with the fungal molecule β-glucan induced epigenetic and transcriptomic rewiring of GMPs and neutrophils that was associated with a type I IFN signature, and inhibition of type I IFN signaling abrogated the enhanced granulocyte responses (Kalafati et al. 2020). In humans, vaccination with tuberculosis vaccine bacillus Calmette-Guérin (BCG) caused long-term epigenetic changes that augmented neutrophils’ antimicrobial functions (Moorlag et al. 2020). However, trained immunity is also induced by endogenous molecules (danger-associated molecular patterns [DAMPs]) and continuous infections and in these settings can be maladaptive, causing chronic inflammation. Chronic inflammation caused by a brief period of experimentally induced periodontitis in mice caused lasting maladaptive trained immunity involving transcriptomic changes in HSPCs, an overresponse of myeloid cells to the TLR4 ligand LPS, and systemic inflammation that increased the severity of arthritis, which is a frequent comorbidity in patients with periodontal disease (Li et al. 2022). The epigenetic changes in humans caused by trained innate immunity persist from 3 mo to 1 yr, with some studies reporting augmented vaccine-induced innate immune responses in children for up to 5 yr (Bekkering et al. 2021). We propose that loss or haploinsufficiency of RUNX1 causes a permanent fixed state of maladaptive innate immunity. Furthermore, we hypothesize that the intrinsic changes in HSPCs and innate immune effector cells likely contribute to the increased incidence of inflammatory and allergic conditions in FPDMM patients and possibly the elevated leukemia risk.
RUNX1 loss in GMPs results in the epigenetic and transcriptional derepression of a subset of retroelements in neutrophils, specifically LINEs and LTRs/ERVs. This is likely caused by elevated tonic type I IFN signaling, based on the enrichment of IRF and STAT1::STAT2 motifs in the chromatin of retroelements that gained accessibility and previous studies showing that LTRs/ERVs (e.g., ERVKs) are regulated by IRFs and STATs (Manghera and Douville 2013). It remains to be determined whether the increased expression of dsRNA from retroelements is only a consequence of elevated tonic type I IFN signaling or whether dsRNA contributes to the response by activating cytosolic sensors that amplify type I IFN signaling in an autocrine or paracrine manner.
Mutations in several genes associated with clonal hematopoiesis of indeterminate potential (CHIP) increase the inflammatory properties of myeloid lineage cells (Jaiswal and Libby 2020). CHIP is caused by somatic mutations acquired by HSCs that confer a selective advantage to the mutated HSC, causing it to preferentially expand in the BM relative to unmutated HSCs (Genovese et al. 2014; Jaiswal et al. 2014). CHIP elevates the risk of cardiometabolic diseases by increasing the inflammatory properties of myeloid lineage cells that differentiate from the mutated HSCs (Jaiswal and Libby 2020). To our knowledge, the derepression of TEs in myeloid lineage cells with CHIP mutations has not been demonstrated, but we believe it is worth examining, particularly as the most common CHIP mutations involve epigenetic regulators (DNMT3A, TET2, and ASXL1) and a kinase in the type II IFN pathway (JAK2) (Jaiswal and Libby 2020).
Our studies focused on neutrophils, but the regulation of type I IFN signaling and chromatin accessibility of retroelements by RUNX1 is not necessarily confined to this lineage. Previous work demonstrated that loss of RUNX1 function in lung alveolar epithelial cells, B cells, and BM cells increased the production of type I IFNs and/or ISGs (DeKelver et al. 2014; Thomsen et al. 2021; Hu et al. 2022). Therefore, FPDMM patients may have a more generalized dysregulation of tonic type I interferon signaling and inflammation involving multiple tissues in which RUNX1 is expressed that could contribute to their inflammatory conditions.
Materials and methods
Mice
Runx1ΔHSC, Runx1ΔLYM, or Runx1ΔGMP mice were created by breeding Runx1f/f (Runx1tm1Spe) mice (Growney et al. 2005) with Vav1-Cre [Tg(Vav1-cre)1Graf] (Stadtfeld and Graf 2005), Rag1-Cre [Rag1tm1(cre)Thr] (McCormack et al. 2003), or Cebpa-Cre [Cebpatm1.1(cre)Touw] (Wolfler et al. 2010) mice. Rag2−/− mice (B6.Cg-Rag2tm1.1Cgn/J) were purchased from JAX. Cd14−/− mice (B6.129S4-Cd14tm1Frm/J) (Moore et al. 2000) were obtained from Carla R. Scanzello. Runx1R188Q/+ (C57BL/6J-Runx1<tm1Lhc>R188Q) knock-in mice are described elsewhere. Male and female mice ages 6–12 wk were used in all experiments. Mice were handled according to protocols approved by the University of Pennsylvania's Institutional Animal Care and Use Committee and housed in a specific pathogen-free facility.
Flow cytometry and cell sorting
A full list of antibodies is provided in Supplemental Table S1. Flow cytometry was performed on an LSR II, and data were analyzed using FlowJo software. The lineage panel in Supplemental Figure S8 includes CD3, CD11b, B220, Gr-1, Nk1.1, and Ter119. Cells analyzed by intracellular flow were fixed and permeabilized using Cytofix/Cytoperm (Benton Dickinson [BD]) prior to intracellular staining in the perm/wash buffer (BD). For dsRNA intracellular flow, cells were permeabilized with 3% PFA for 15 min on ice, washed twice with FACS buffer (2% FBS in 1× PBS), and then permeabilized with 0.1% Saponin for 15 min on ice. The cells were then incubated with 9D5 dsRNA rabbit IgG mAb (1:500) in 0.1% Saponin for 15 min on ice, washed twice with 0.1% Saponin, and then incubated with donkey antirabbit IgG (minimal x-reactivity) PE antibody (1:400) for 15 min on ice. The cells were rinsed three times with FACS buffer. Gating schemes for intracellular flow assays are provided in Supplemental Figures S1A and S8. A BD fluorescence-activated cell sorter (FACS) Aria II was used to sort cells at 482.63 kPa (70 psi) using a 70-μm nozzle. Gating schemes for purified neutrophils and GMPs are provided (Supplemental Fig. S8).
Statistical analysis
Unless otherwise indicated, all statistical analyses were performed using Prism/GraphPad/R.
FPDMM patient data
FPDMM patient data were obtained following informed consent under the clinical study entitled “Longitudinal Studies of Patients with FPDMM” (ClinicalTrials.gov identifier NCT03854318).
Data and code availability
The data discussed here have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO series accession number GSE221427 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221427).
Supplementary Material
Acknowledgments
We thank William Murphy, Jennifer Jakubowski, and Shifu Tian in the Flow Cytometry and Cell Sorting Resource Laboratory for cell sorting assistance. We thank Carla Scanzello for the Cd14−/− mice, and Andrew Modzelewski for helpful advice. We especially thank Boyoung Shin and Ellen Rothenberg for sharing their CUT&RUN protocol. This work was supported by R01HL091724 and U01HL100405 to N.A.S., 1F30DK128926-01A1 and T32HL007439 to A.U.Z., 1F31HL150952-01 and T32 HD083185 to E.D.H., the RUNX1 Research Program (N.A.S.), and the Intramural Research Program, National Human Genome Research Institute, National Institutes of Health (E.B., J.D., and P.P.L.).
Author contributions: N.A.S., A.U.Z., and W.Y. conceived the project and wrote the manuscript. All authors discussed the results and revised the manuscript. A.U.Z., D. Yen, E.D.H., D. Ye, J.-g.R., and J.D. performed the experiments. W.Y., D. Yen, and A.U.Z. generated the genomics data. W.Y., A.U.Z., and E.D.H. performed bioinformatics analyses. M.H.A, L.H.C., and I.P.T. provided mice. A.J.M, W.T., and K.T. provided advice. P.P.L., E.B., and J.D. provided FPDMM patient samples.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Aritcle and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350418.123.
Competing interest statement
The authors declare no competing interests.
References
- Bekkering S, Domínguez-Andrés J, Joosten LAB, Riksen NP, Netea MG. 2021. Trained immunity: reprogramming innate immunity in health and disease. Annu Rev Immunol 39: 667–693. 10.1146/annurev-immunol-102119-073855 [DOI] [PubMed] [Google Scholar]
- Bellissimo DC, Speck NA. 2017. RUNX1 mutations in inherited and sporadic leukemia. Front Cell Dev Biol 5: 111. 10.3389/fcell.2017.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissimo DC, Chen CH, Zhu Q, Bagga S, Lee CT, He B, Wertheim GB, Jordan M, Tan K, Worthen GS, et al. 2020. Runx1 negatively regulates inflammatory cytokine production by neutrophils in response to Toll-like receptor signaling. Blood Adv 4: 1145–1158. 10.1182/bloodadvances.2019000785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Arts P, Carmichael CL, Babic M, Dobbins J, Chong CE, Schreiber AW, Feng J, Phillips K, Wang PPS, et al. 2020. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv 4: 1131–1144. 10.1182/bloodadvances.2019000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Hur S. 2022. Cellular origins of dsRNA, their recognition and consequences. Nat Rev Mol Cell Biol 23: 286–301. 10.1038/s41580-021-00430-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churpek JE, Pyrtel K, Kanchi KL, Shao J, Koboldt D, Miller CA, Shen D, Fulton R, O'Laughlin M, Fronick C, et al. 2015. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood 126: 2484–2490. 10.1182/blood-2015-04-641100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielska A, Matyjek M, Kwiatkowska K. 2021. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci 78: 1233–1261. 10.1007/s00018-020-03656-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AR, Triche T Jr, Ramsingh G. 2018. Transposable element expression in acute myeloid leukemia transcriptome and prognosis. Sci Rep 8: 16449. 10.1038/s41598-018-34189-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKelver RC, Lewin B, Weng S, Yan M, Biggs J, Zhang DE. 2014. RUNX1-ETO induces a type I interferon response which negatively effects t(8;21)-induced increased self-renewal and leukemia development. Leuk Lymphoma 55: 884–891. 10.3109/10428194.2013.815351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuitch N, Broadbridge E, Cunningham L, Liu PP. 2021. RUNX1 familial platelet disorder with associated myeloid malignancies. In GeneReviews (ed. Adam MP, et al. ), https://www.ncbi.nlm.nih.gov/books/NBK568319. University of Washington, Seattle. [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinet E, Gu Z, Imbusch CD, Giese NA, Büscher M, Safavi M, Weisenburger S, Klein C, Vogel V, Falcone M, et al. 2021. Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell of origin. Cancer Discov 11: 638–659. 10.1158/2159-8290.CD-20-1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gázquez-Gutiérrez A, Witteveldt J, Heras SR, Macias S. 2021. Sensing of transposable elements by the antiviral innate immune system. RNA 27: 735–752. 10.1261/rna.078721.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis FK, Goff SP. 2020. Silencing and transcriptional regulation of endogenous retroviruses: an overview. Viruses 12: 884. 10.3390/v12080884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371: 2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, et al. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106: 494–504. 10.1182/blood-2004-08-3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan CC, King-Smith SL, Lawrence DM, Arts P, Feng J, Andrews J, Armstrong M, Ha T, Dobbins J, Drazer MW, et al. 2021. The RUNX1 database (RUNX1db): establishment of an expert curated RUNX1 registry and genomics database as a public resource for familial platelet disorder with myeloid malignancy. Haematologica 106: 3004–3007. 10.3324/haematol.2021.278762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Pan Q, Zhou K, Ling Y, Wang H, Li Y. 2022. RUNX1 inhibits the antiviral immune response against influenza A virus through attenuating type I interferon signaling. Virol J 19: 39. 10.1186/s12985-022-01764-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Libby P. 2020. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol 17: 137–144. 10.1038/s41569-019-0247-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371: 2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen JP. 1993. The Runt-domain identifies a new family of heteromeric DNA-binding transcriptional regulatory proteins. Trends Genet 9: 338–341. 10.1016/0168-9525(93)90026-E [DOI] [PubMed] [Google Scholar]
- Kalafati L, Kourtzelis I, Schulte-Schrepping J, Li X, Hatzioannou A, Grinenko T, Hagag E, Sinha A, Has C, Dietz S, et al. 2020. Innate immune training of granulopoiesis promotes anti-tumor activity. Cell 183: 771–785.e12. 10.1016/j.cell.2020.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafati L, Hatzioannou A, Hajishengallis G, Chavakis T. 2022. The role of neutrophils in trained immunity. Immunol Rev 314: 142–157. 10.1111/imr.13142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Chen L-F, Ogawa E, Kim W-Y, Ito Y. 1998. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor a subunit revealed in the presence of the b subunit. Mol Cell Biol 18: 2444–2454. 10.1128/MCB.18.5.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsou K, Lagiou P, Magiorkinis G. 2022. Human endogenous retroviruses in cancer: oncogenesis mechanisms and clinical implications. J Med Virol 95: e28350. 10.1002/jmv.28350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouroukli O, Symeonidis A, Foukas P, Maragkou MK, Kourea EP. 2022. Bone marrow immune microenvironment in myelodysplastic syndromes. Cancers 14: 5656. 10.3390/cancers14225656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson HA, Liang Y, Wang T. 2023. Transposable elements in mammalian chromatin organization. Nat Rev Genet. 10.1038/s41576-023-00609-6 [DOI] [PubMed] [Google Scholar]
- Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. 2019. Identification of transcription factor binding sites using ATAC-seq. Genome Biol 20: 45. 10.1186/s13059-019-1642-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang H, Yu X, Saha G, Kalafati L, Ioannidis C, Mitroulis I, Netea MG, Chavakis T, Hajishengallis G. 2022. Maladaptive innate immune training of myelopoiesis links inflammatory comorbidities. Cell 185: 1709–1727 e18. 10.1016/j.cell.2022.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Zhou SY, Feng DY, Xiao J, Li WY, Xu CD, Wang HY, Zhou T. 2016. Runt-related transcription factor 1 (RUNX1) binds to p50 in macrophages and enhances TLR4-triggered inflammation and septic shock. J Biol Chem 291: 22011–22020. 10.1074/jbc.M116.715953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghera M, Douville RN. 2013. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10: 16. 10.1186/1742-4690-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteucci C, Balestrieri E, Argaw-Denboba A, Sinibaldi-Vallebona P. 2018. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol 53: 17–30. 10.1016/j.semcancer.2018.10.001 [DOI] [PubMed] [Google Scholar]
- McCormack MP, Forster A, Drynan L, Pannell R, Rabbitts TH. 2003. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol Cell Biol 23: 9003–9013. 10.1128/MCB.23.24.9003-9013.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. 2018. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front Immunol 9: 1135. 10.3389/fimmu.2018.01135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Andersson LP, Ingalls RR, Monks BG, Li R, Arnaout MA, Golenbock DT, Freeman MW. 2000. Divergent response to LPS and bacteria in CD14-deficient murine macrophages. J Immunol 165: 4272–4280. 10.4049/jimmunol.165.8.4272 [DOI] [PubMed] [Google Scholar]
- Moorlag S, Rodriguez-Rosales YA, Gillard J, Fanucchi S, Theunissen K, Novakovic B, de Bont CM, Negishi Y, Fok ET, Kalafati L, et al. 2020. BCG vaccination induces long-term functional reprogramming of human neutrophils. Cell Rep 33: 108387. 10.1016/j.celrep.2020.108387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. 2007. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446: 685–689. 10.1038/nature05673 [DOI] [PubMed] [Google Scholar]
- Perera PY, Vogel SN, Detore GR, Haziot A, Goyert SM. 1997. CD14-dependent and CD14-independent signaling pathways in murine macrophages from normal and CD14 knockout mice stimulated with lipopolysaccharide or taxol. J Immunol 158: 4422–4429. 10.4049/jimmunol.158.9.4422 [DOI] [PubMed] [Google Scholar]
- Platanitis E, Demiroz D, Schneller A, Fischer K, Capelle C, Hartl M, Gossenreiter T, Müller M, Novatchkova M, Decker T. 2019. A molecular switch from STAT2–IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat Commun 10: 2921. 10.1038/s41467-019-10970-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco K, Laky K, Li M, Merguerian M, Craft K, Cunningham L, Liu P, Guerrerio P. 2020. Germline RUNX1 deficiency predisposes to allergy and autoimmunity. J Allergy Clin Immunol 147: AB68. 10.1016/j.jaci.2020.12.266 [DOI] [Google Scholar]
- Saleh A, Macia A, Muotri AR. 2019. Transposable elements, inflammation, and neurological disease. Front Neurol 10: 894. 10.3389/fneur.2019.00894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68: 855–867. 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6: e21856. 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son KN, Liang Z, Lipton HL. 2015. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J Virol 89: 9383–9392. 10.1128/JVI.01299-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-J, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, et al. 1999. Haploinsufficiency of CBFA2 (AML1) causes familial thrombocytopenia with propensity to develop acute myelogenous leukamia (FPD/AML). Nature Genet 23: 166–175. 10.1038/13793 [DOI] [PubMed] [Google Scholar]
- Sorrell A, Espenschied C, Wang W, Weitzel J, Chu S, Parker P, Saldivar S, Bhatia R. 2012. Hereditary leukemia due to rare RUNX1c splice variant (L472X) presents with eczematous phenotype. Int J Clin Med 3: 607–613. 10.4236/ijcm.2012.37110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Graf T. 2005. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development 132: 203–213. 10.1242/dev.01558 [DOI] [PubMed] [Google Scholar]
- Stubbins RJ, Platzbecker U, Karsan A. 2022. Inflammation and myeloid malignancy: quenching the flame. Blood 140: 1067–1074. 10.1182/blood.2021015162 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kusuda M, Yamaguchi Y. 2022. Interferons and systemic lupus erythematosus: pathogenesis, clinical features and treatments in interferon-driven disease. Mod Rheumatol 10.1093/mr/roac140 [DOI] [PubMed] [Google Scholar]
- Tang X, Sun L, Jin X, Chen Y, Zhu H, Liang Y, Wu Q, Han X, Liang J, Liu X, et al. 2017. Runt-related transcription factor 1 regulates LPS-induced acute lung injury via NF-κB signaling. Am J Respir Cell Mol Biol 57: 174–183. 10.1165/rcmb.2016-0319OC [DOI] [PubMed] [Google Scholar]
- Thomsen I, Kunowska N, de Souza R, Moody AM, Crawford G, Wang YF, Khadayate S, Whilding C, Strid J, Karimi MM, et al. 2021. RUNX1 regulates a transcription program that affects the dynamics of cell cycle entry of naive resting B cells. J Immunol 207: 2976–2991. 10.4049/jimmunol.2001367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yin Y, Xu L, Su J, Huang F, Wang Y, Boor PPC, Chen K, Wang W, Cao W, et al. 2017. Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections. Sci Signal 10: eaah4248. 10.1126/scisignal.aah4248 [DOI] [PubMed] [Google Scholar]
- Wolfler A, Danen-van Oorschot AA, Haanstra JR, Valkhof M, Bodner C, Vroegindeweij E, van Strien P, Novak A, Cupedo T, Touw IP. 2010. Lineage-instructive function of C/EBPα in multipotent hematopoietic cells and early thymic progenitors. Blood 116: 4116–4125. 10.1182/blood-2010-03-275404 [DOI] [PubMed] [Google Scholar]
- Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. 2008. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol 9: 378–387. 10.1038/ni1576 [DOI] [PubMed] [Google Scholar]
- Zanoni I, Granucci F. 2013. Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol 3: 32. 10.3389/fcimb.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed here have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO series accession number GSE221427 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221427).