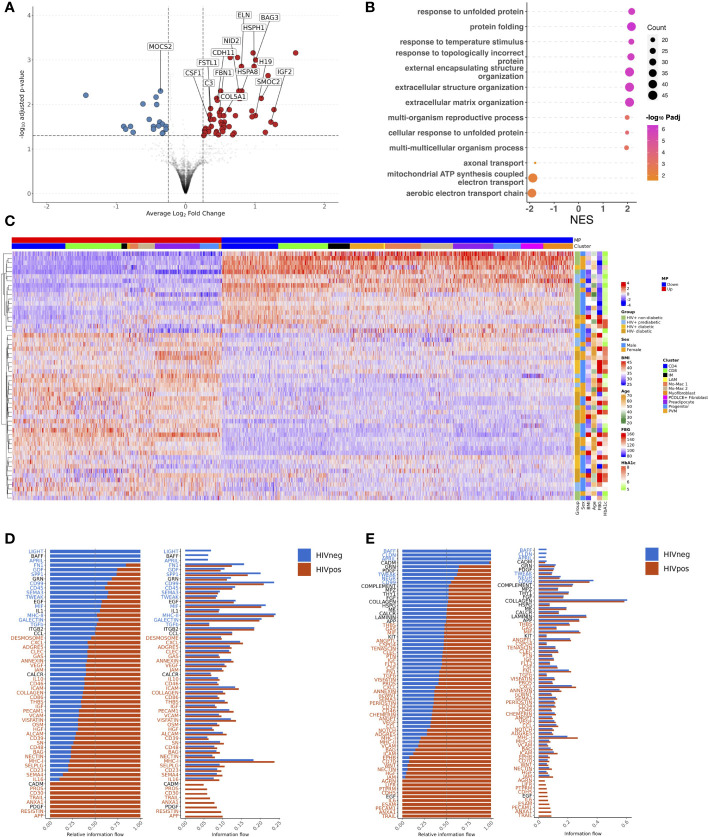
Figure 10.
HIV-negative diabetic and HIV-positive diabetic have a similar multicellular gene expression program but divergent intercellular communication pathways. (A) Preadipocyte volcano plot with average Log2 fold change (x-axis) and –log10 adjusted p-value (y-axis) for HIV-positive diabetic vs HIV- diabetic (reference) persons. Genes that had ≥ 0.25 log2 fold change and adjusted p-value < 0.05 were colored red (higher expression) and blue (lower expression). (B) Gene set enrichment analysis (GSEA) using the Gene Ontology database. The top over and under enriched pathways were included with normalized enrichment score (NES) on x-axis and descriptive term on y-axis. Dot size represents the number of gene hits in the pathway and dot color represents the –log10 adjusted p-value. (C) Average scaled expression of top genes from all cells in Multicellular Program (MP) 1 sorted by expression (columns), across samples plotted with hierarchical clustering (rows) and labeled with clinical variables including body mass index (BMI), age, sex, and measures of glucose intolerance. (D, E) Bar plot with relative information flow (left) or overall information flow (right) on the x-axis and predicted ligand-ligand receptor pathways from source cells to target cells. Signaling in HIV+ diabetic is shown in orange while signaling in HIV- diabetic is shown in blue. Pathways with significantly greater interaction in HIV+ diabetics based on Wilcoxon rank sum are labeled in orange while pathways with significantly greater interaction in HIV- diabetics are labeled in blue (p < 0.05). (D) Macrophages (source) to all cells (target). (E) Stromal (source) to all cells (target). BMI, body mass index; FBG, fasting blood glucose; HbA1c, hemoglobin A1c; IM, intermediate macrophage; LAM, lipid-associated macrophage; NES, normalized enrichment score; PVM, perivascular macrophage.