Fig. 2: Tuning the tensile strength of sNRR domains.
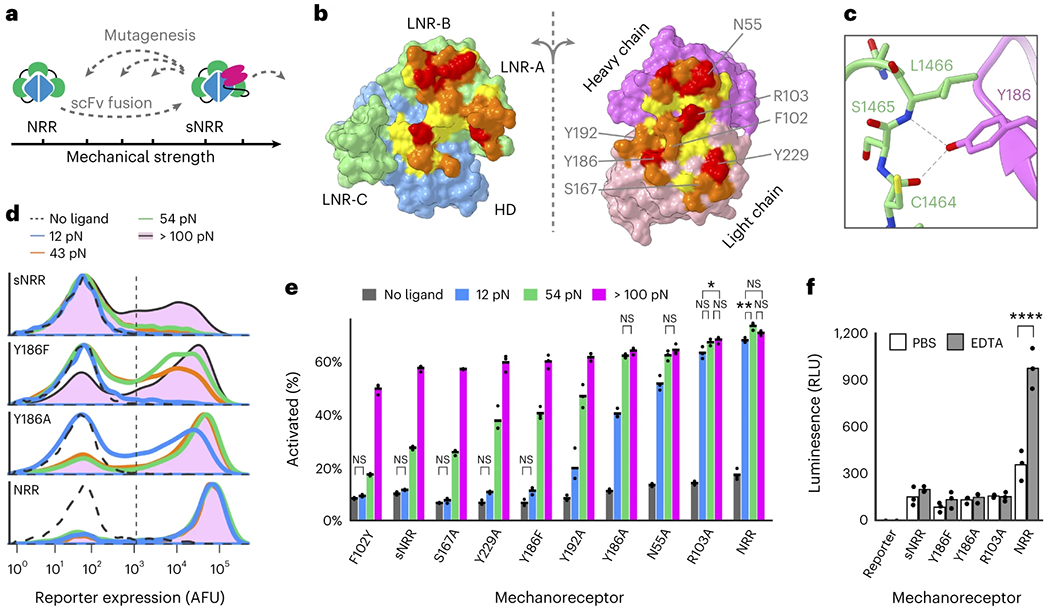
a, Schematic of strategy to tune mechanical strength. b, Open-book view of the interface between the Notch1 NRR and the NRR-binding Fab. The HD and LNRs of the NRR are blue and green, respectively, and the heavy and light chains of the scFv are magenta and pink. Additional residue coloring corresponds to the distance between the two surfaces (<5.5 Å, yellow; 5.5–4 Å, orange; 4–3 Å, red) (PDB code: 3L95). c, Y186 on the Fab light chain is highlighted as an example of a mutated residue. d, Mutating Y186 decreases sNRR mechanical strength. Receptors stably integrated into HEK293-FT cells upregulate the expression of a UAS-driven H2B-mCherry reporter gene upon activation by TGTs. Traces represent normalized densities of total cells within transduced populations from three independent experiments (n = 3). e, Mutating sNRR residues creates receptors with a range of force activation thresholds. Receptors are transiently expressed in HEK293-FT cells containing a stably integrated UAS-H2B-mCherry reporter gene. Mean from three independent transfections (n = 3) displayed, quantified by flow cytometry (over 5,000 cells assessed per replicate) and analyzed with two-way ANOVA (mechanosensor and TGT threshold). All unlabeled comparisons within the same mechanosensor have P < 0.0001; labeled NS, P > 0.05, *P < 0.05, **P < 0.01, otherwise. f, Cells expressing SynNotch receptors with an NRR domain or sNRR domain of various strengths are treated with either 0.5 mM EDTA or PBS for 15 min at 37 °C followed by the re-addition of culture media. Reporter expression was evaluated 6 h later. Activated receptors upregulate the expression of a UAS-regulated luciferase reporter gene. Mean from three independent transfections (n = 3), two-way ANOVA, P value labeled **** for P < 0.0001, all unlabeled comparisons have P > 0.05.
