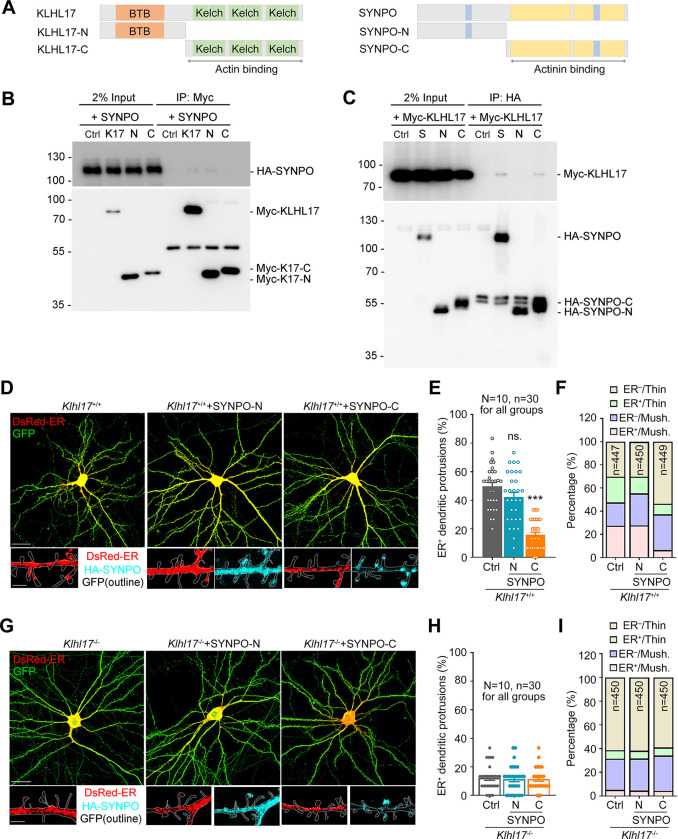
Fig 8. KLHL17 and SYNPO association controls the dendritic spine distribution of the ER.
(A) Schematic domain structures of KLHL17 and SYNPO. Left: Full-length KLHL17 and truncated KLHL17-N (amino acid residues 1–300) and KLHL17-C (amino acid residues 301–640) mutants. KLHL17-N contains the BTB domain, whereas KLHL17-C comprises 6 Kelch domains. Right: Full-length SYNPO and truncated SYNPO-N (amino acid residues 1–299) and SYNPO-C (amino acid residues 300–690) mutants. SYNPO-C includes 2 regions required for the actinin-binding domain (yellow blocks) and one 14-3-3 binding motif (blue block). The other 14-3-3 binding motif is located in the N-terminal region of SYNPO. (B, C) C-terminal SYNPO (SYNPO-C) co-immunoprecipitates with N-terminal KLHL17 (KLHL17-N). Neuro-2A cells were cotransfected with either full-length Myc-KLHL17 or HA-SYNPO with the N- or C-terminal constructs, as indicated, and subjected to immunoprecipitation and immunoblotting using the indicated antibodies. (D–F) Overexpression of SYNPO-C reduces the number of ER-positive dendritic spines of wild-type neurons. Cultured neurons were transfected with the indicated plasmids at 12 DIV and harvested for immunostaining at 18 DIV. (D) Representative images of the whole cells and enlarged dendrite segments. SYNPO-C exhibits a punctate signal like that of full-length SYNPO, whereas SYNPO-N shows a diffuse pattern. (E) Quantification of the percentage of ER+ dendritic spines. (F) Quantification of the percentage of ER+ protrusions in thin and mushroom-like (Mush.) spines. (G–I) The same experiments as (D–F), except that Klhl17–/–neurons were used. In (E) and (H), the sample sizes of examined neurons (N) and dendritic segments (n) are indicated. In (F) and (I), the numbers of dendritic spines (n) examined are indicated. All samples were collected from 2 independent experiments. The data represent mean ± SEM. Individual data points are also shown. *** P < 0.001; ns, not significant; one-way ANOVA. Scale bars: (D), (G) whole cells: 20 μm; enlarged segment: 2 μm. The numerical value data and statistical results are available in S1 and S2 Data, respectively. BTB, Bric-a-brac/Tramtrack/Broad; DIV, day in vitro; ER, endoplasmic reticulum; KLHL17, Kelch-like protein 17.