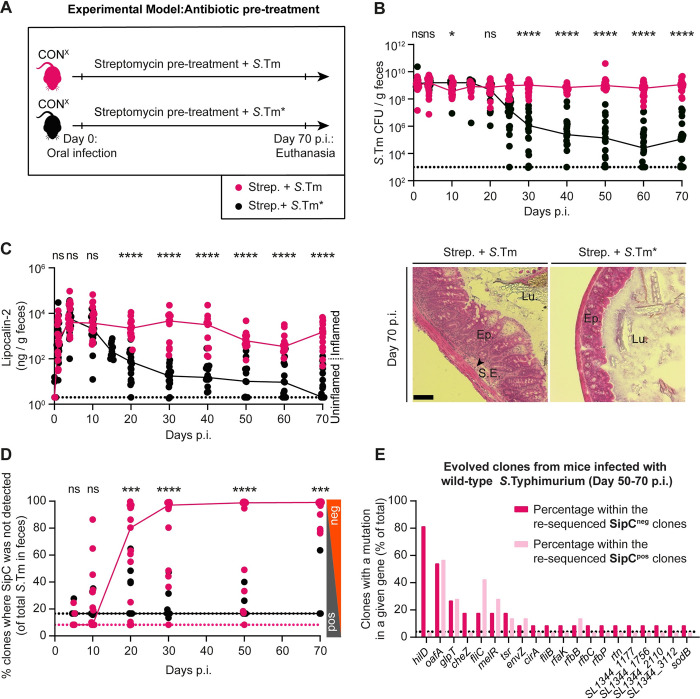
Fig 1. Selection for mutants with reduced virulence in streptomycin pretreated CONX mice infected with wild-type S.
Typhimurium or S.Tm*. (A) Scheme summarizing the experiment in Panels B–D. (B–D) Streptomycin pretreated CONX mice were infected with wild-type S. Typhimurium (SL1344 WT; pink, filled circles; n = 19; 4 independent experiments) or S.Tm* (SL1344 ΔssaV; black, filled circles; n = 22; 3 independent experiments) for 70 days. (B) Fecal Salmonella populations as determined using MacConkey plates with selective antibiotics. (C) Gut inflammation. Left: ELISA data measuring the Lipocalin-2 concentration in fecal pellets. We analyzed samples from at least n = 11 animals per time point. Dotted lines indicate the detection limit. Colored lines connect the medians. Right: representative images of HE-stained cecum tissue sections showing intestinal crypts to assess the severity of enteric disease [19]; scale bar 100 μm. (D) Percentage of colonies without detectable SipC (as measured by colony protein blot). Colored lines connect the medians. Two-tailed Mann–Whitney U tests were used to compare the wild-type S. Typhimurium to the S.Tm* data (p ≥ 0.05 not significant (ns), p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****)). Source data can be found in S1 Data file. (E) Whole-genome sequencing was performed from clones re-isolated from mice in Panels B–D. A complete overview of non-synonymous mutations is summarized in S1–S6 Tables. The graph illustrates the 20 genes most frequently mutated in clones re-isolated from mice infected with wild-type S. Typhimurium at day 50–70 p.i. Dark pink: mutations from clones without detectable SipC expression (n = 11 independent clones were analyzed). Light pink: mutations from clones where SipC was detected (n = 11 independent clones were analyzed). The dotted line indicates the percentage that corresponds to a mutation which occurs in just 1 clone. Genes mutated only in mutS mutant strains (i.e., mutator clones) are excluded. Only non-synonymous mutations and genes disrupted by stop codons or frameshifts are shown.